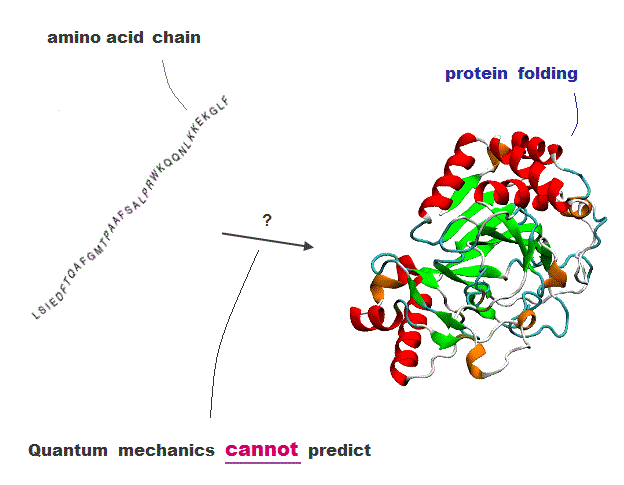
(Fig.1) Amino acid chain → protein folding = cannot be predicted !

The current methods cannot predict how amino acid chains fold into proteins.
Because quantum mechanics is so old that it cannot handle multi-electron atoms.
(Fig.2) Atomic bonds inside proteins rotate almost freely → many forms !
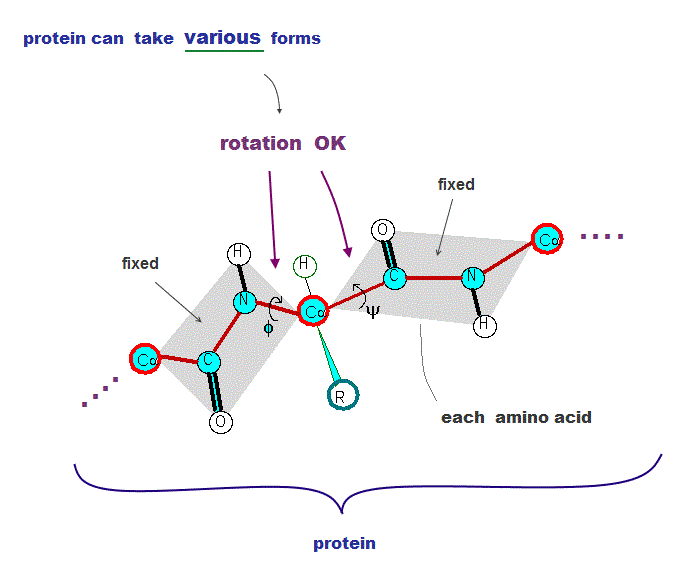
All proteins consist of amino acids. The point is all atomic bonds except peptide bonds can rotate freely inside proteins.
So each amino acid chain can take almost infinite kinds of different protein structures under the same sequence.
(Fig.3) Protein chain like a rope can take many different structures.
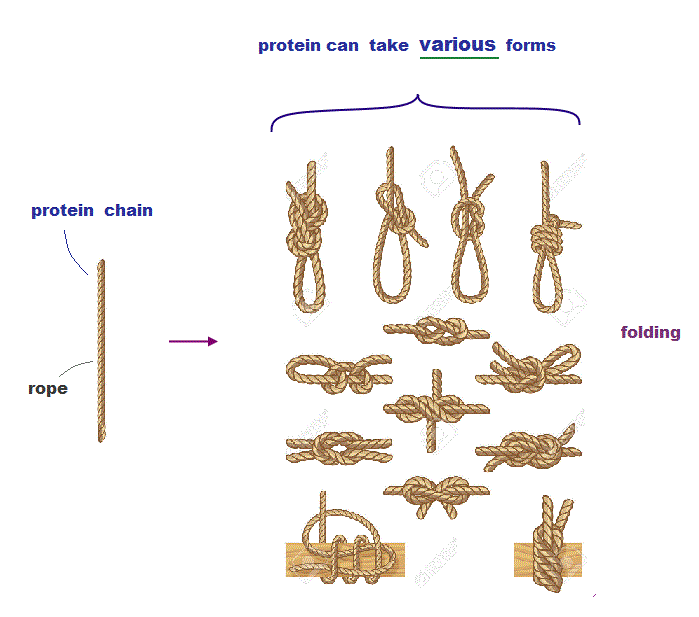
Protein chain resembles a rope which can form many different knots from the same rope (= amino acid chain ).
Each different knot is stable. So even if each amino acid chain takes different protein structure, it can also remain stable in energy.
This is one of the main reasons why the current method ( pursuing only the 'single' lowest energy state ) cannot predict protein structure.
Different stable knots above correspond to 'local energy minima'. There are almost infinite local energy minima in protein folding.
(Fig.4) Two H2O molecules cannot approach less than 1.8 Å ← resistance !
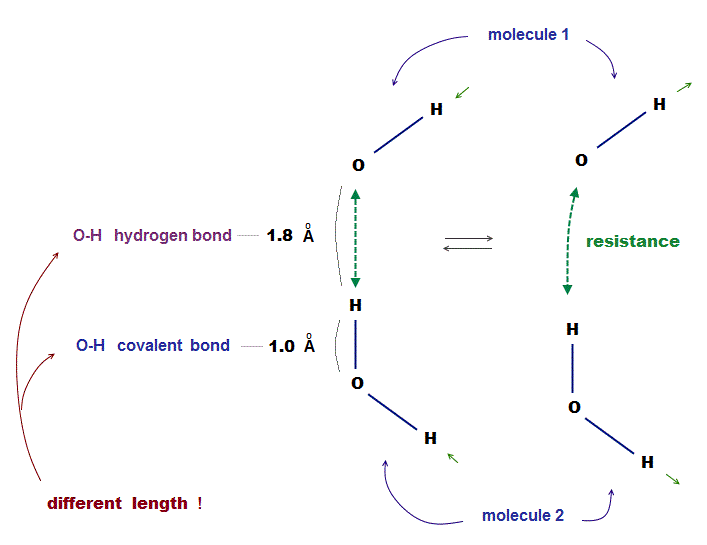
Each atomic bond length is specific and almost fixed under the common physical law. This also applies to non-covalent bonds such as hydrogen bond.
The point is each molecule cannot approach each other closer than some critical length ( O-H > 1.8 Å ), which is specific to each atom.
When two molecules are closer to each other than this 'critical point', some extremely strong "repulsive force" prevents them, like "resistance" between two things.
The current quantum mechanics cannot describe this "resistance" in non-covalent bond using "concrete" objects, which makes it impossible to predict protein structure.
(Fig.5) ↓ If there are other obstacles, protein is folded "differently"
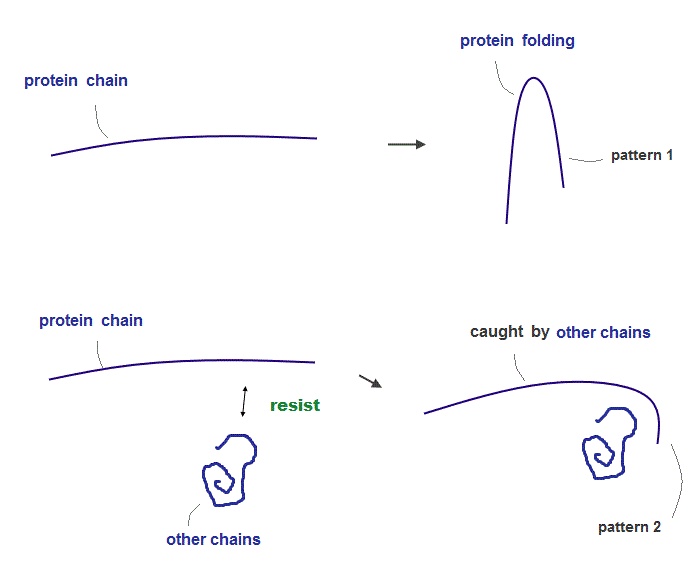
The existence of strong "resistance" between atoms and molecules play a crucial role in determining final protein structures.
When there are some other obstacles (= amino acid chain ), a protein chain is folded differently.
This "resistance" is so strong that no molecules cannot be closer to each other than some critical point ( they can be farther away from the critical point ).
Using PDB protein data, you can find that any non-covalent bond length between O (or N ) and H cannot be shorter than about 1.8 Å due to this strong resistance.
(Fig.6) Final protein structures change depending on "folding process" !
If two atomic chains cannot be closer to each other than some 'critical point', these strong repulsions (= resistance ) determine the final protein structure like a rope.
It means final protein structures change, depending on "folding process" ( using enzymes ) rather than their amino acid sequences.
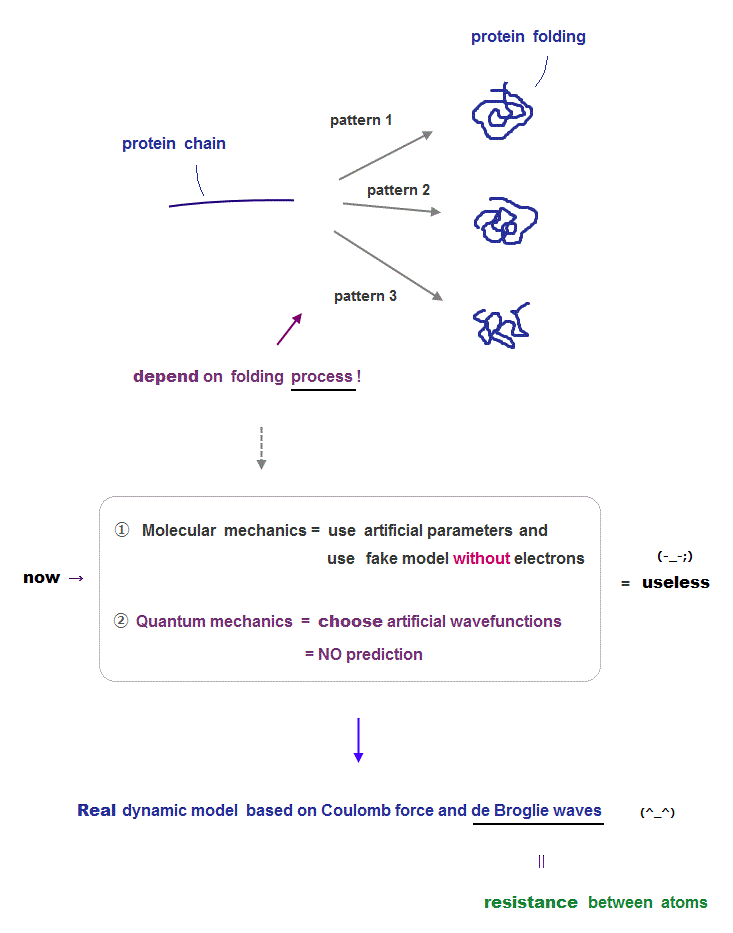
Now there are two computational methods for protein structures: molecular mechanics and old quantum mechanics.
Molecular mechanics use artificial classical models with artificial parameters which can be manipulated freely, so it has No ability to predict unkown protein structure.
They try to seek only the lowest energy state using those artificial parameters adjusted by the existing experimental data. But this cannot predict final protein structure.
Because depending on the folding process, the final stable structures change (= one of which corresponds to energy 'local minima' ).
Quantum mechanics just choose fake wavefunction out of infinite choices, so it's also impossible to pedict unknown new protein structures.
(Fig.7) Parameters (= bond length, angle, torsion ) are adjusted artificially.
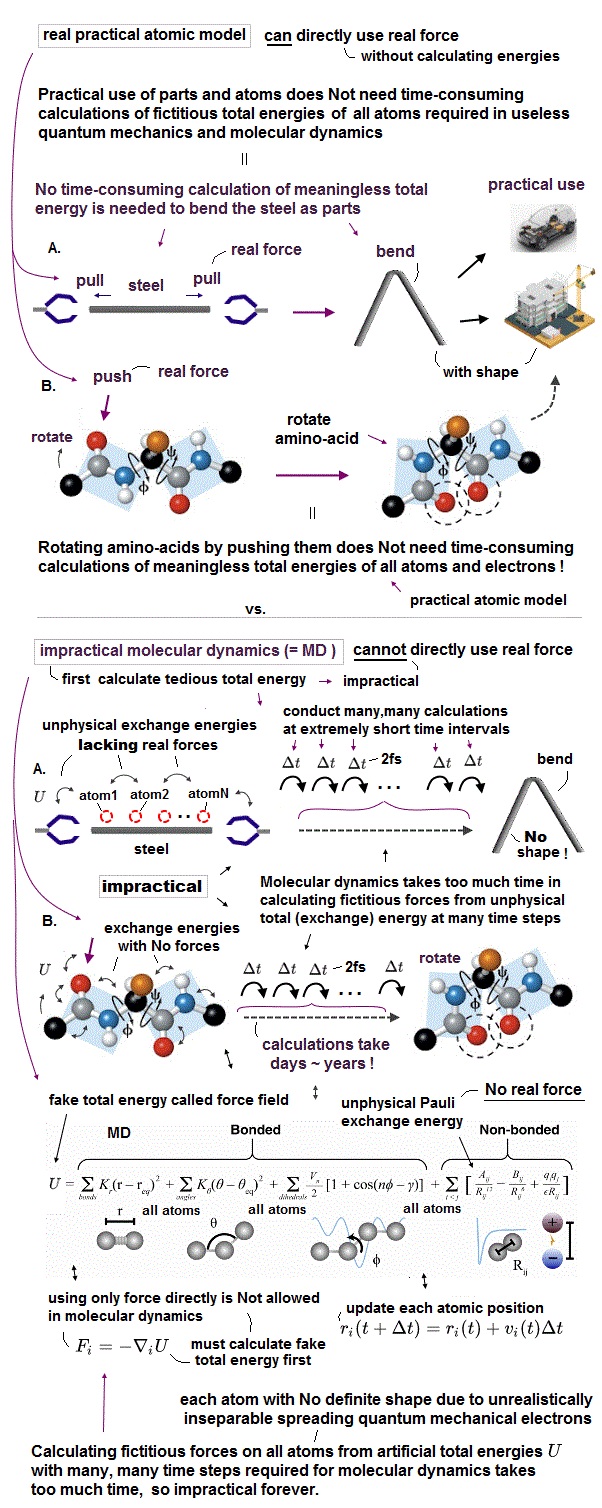
They often use molecular mechanics which uses fake "classical model" to calculate large molecules and proteins, which quantum mechanics cannot handle.
In fact, this molecular mechanics cannot predict any new physical values, because its model must rely on existing experimental values (= empirical ).
So this model's parameters must be adjusted in advance to fit experimental data. Molecular mochanics cannot be used to predict new chemical reactions.
Even if you choose some bond ( torsion ) angle parameters from other protein data, you cannot predict new unknown protein structure.
Because the final protein structure changes depending on the "protein folding process" due to strong resistence between atoms.
Furthermore, this artificial classical model don't use actual electrons, so it cannot tell us the process of how protein folds, interacting with enzymes.
(Fig.8) Multi-electron Couloum energy → fake potential in DFT, so meagninelss
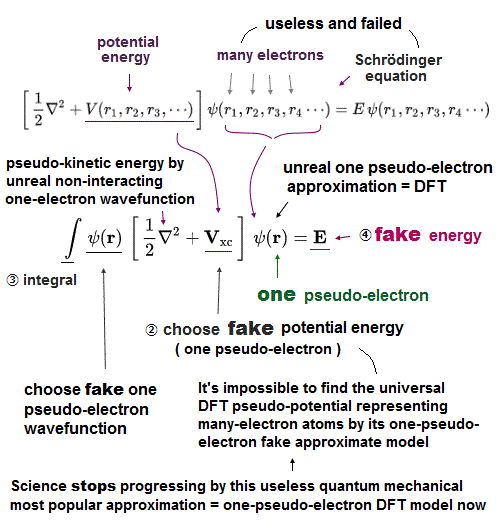
The above molecular mechanics uses fake classical model without any actual electrons.
So they have to rely on old quantum mechanics to predict new chemical reactions among proteins and enzymes.
The problem is this Schrödinger equation has NO exact solutions in multi-electron atoms, so they just artificially pick up fake solutions (= basis set wavefunction ).
Also in density functional theory, they just choose 'fake potential' out of infinite choices.
So all these quantum mechanical methods have NO ability to predict unknown protein structures.
(Fig.9) ↓ Carbon can bind to another carbon ( NOT Neon ). Why ?
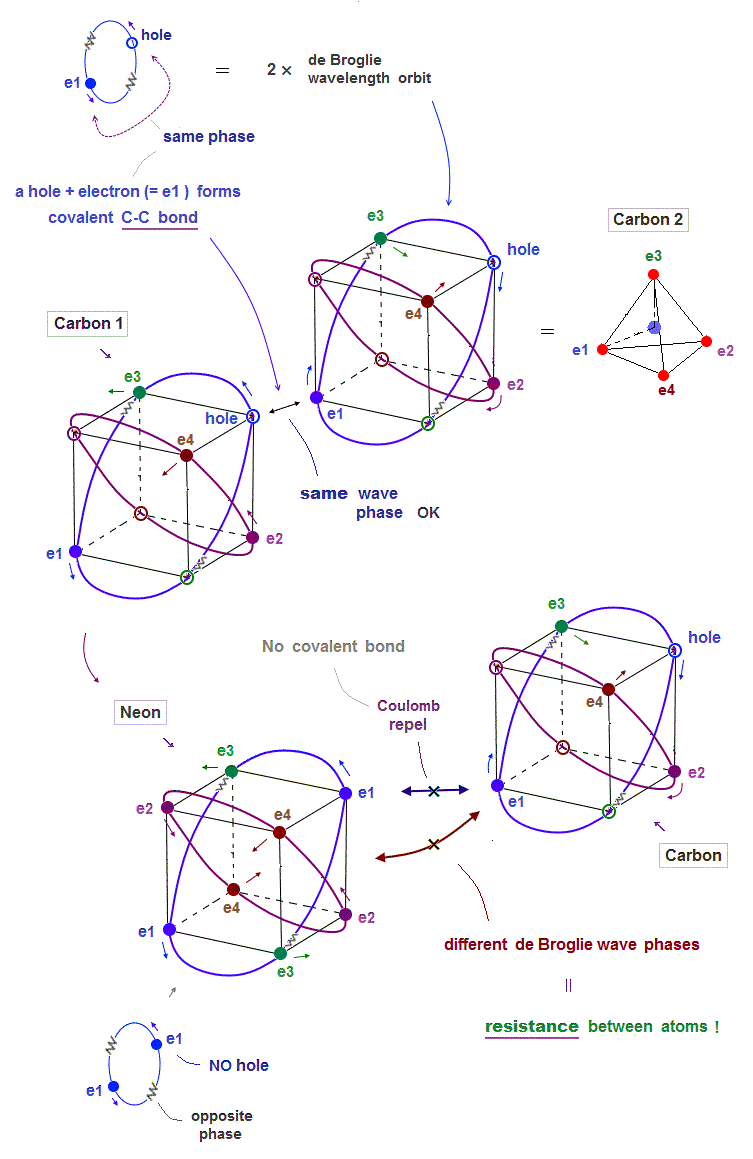
The current quantum mechanics with unrealistic electron spin cannot explain strong atomic "resistance" such as Pauli exclusion principle.
There is the only "de Broglie wave interference" ( and Coulomb force ) which can cause this strong repulsions and molecular bond length.
This can explain real atomic structure where a pair of a hole and electron forms covalent molecular bond such as carbon-carbon ( C-C ) bond.
But Neon cannot form molecular bond with carbon. Why ?
Too weak electron spin cannot explain it.
An electron of carbon tries to avoid Neon's electron due to Coulomb repulsions. There are NO holes (= the same wave phase as electron place ) in de Broglie wave in Neon.
So this Carbon electron has No choice but to approach Neon's different phases of electron's de Broglie wave, which cause them destructive interference.
So Coulomb repulsions and "destructive interference of de Broglie waves" between two electrons cause strong resistance between two atoms !
(Fig.10) We can use atomic resistance as atomic "hook" or "tweezers"
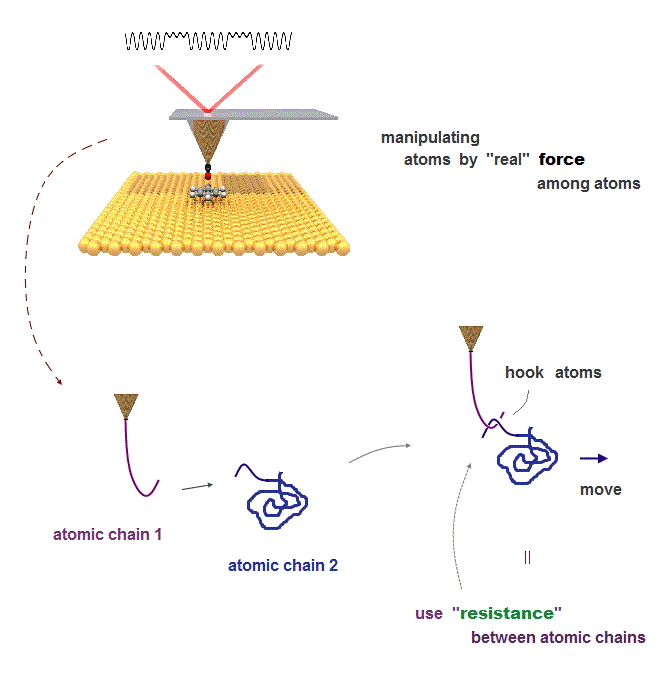
If we define and use this "real" resistance between two atoms ( or molecules ) as a tool, we can manipulate each atom ( or atomic chain ) freely.
For manipulation, the technique of moving each atoms freely is indispensable.
If there are real resistance between atoms, we can use atom as if they were "actual things" such as tweezers or hook to carry other atoms.
The current basic science is shackled to the old Schrödinger equation which has obstinately avoided admitting the existence and definition of "real concrete force".
Because the moment they try to investigate the cause of force, they find all quantum mechanical concepts are unrealistic and wrong.

2017/2/17 updated. Feel free to link to this site.