Home page
X-ray, Cryo-EM are useless
(Fig.1) Today's useless atomic force (or scanning tunneling ) microscope with just one probe tip can only see (useless) planar flat molecules. Only multi-probe microscope can observe and manipulate various 3-dimensional (= 3D ) molecules.

Since 1980s, we have been able to see and manipulate single atoms by atomic force microscopes (= AFM ).
Other cryo-electron microscopes or X-ray crystallography can only vaguely detect electrons and X-rays randomly scattered from proteins, cannot see individual atoms due to their too bad resolution ( this-Limitations, this-3rd,7th-paragraphs ).
This-9th~10th-paragraphs say -- Bad electron microscope
"However, cryo-EM (= cryo electron microscope ) is ineffective for imaging very small proteins.. the technique requires very high sample homogeneity, which increases the difficulties of obtaining high-resolution images of flexible proteins. Moreover, the current resolution of cryo-EM is Not sufficient for pharmaceutical research and development, as the images obtained by this technique have a low signal-to-noise ratio in certain instances."
So only atomic force microscope has potential to clarify atomic mechanisms of proteins, cells.. to cure the current intractable diseases.
Surprisingly, atomic force microscopes (= AFM ) have had only one useless probe tips for 40 years with No progress ( this-Figure.1 ).
The atomic force microscope with only one probe can see only flat, non-3-dimensional molecules such as benzenes ( this-p.4, this or this-p.12-3D molecules, this or this-lower-challenge ), or the upper part of a molecule ( this-p.4-p.5, this-p.2 ).
Today's useless atomic force microscope with only one probe tip can see only flat molecules, because a 3-dimensional (= 3D ) molecule is unstably moved or rotated by being pushed by the microscope probe tip ( this-p.4 ).
So today's useless 3D atomic force microscopes try to see only macroscopic objects or near-flat molecules by dull probe tips without seeing a 3-dimensional molecule ( this-p.3 = this, this-Fig.1 ).
Only atomic force microscopes with multiple probes can see and manipulate any 3-dimensional molecules and proteins to clarify atomic mechanisms and cure diseases.
↑ While one probe fixes a target molecule, other probes can touch and investigate the molecule to clarify atomic structures, which is impossible in the present atomic force microscope with only one useless probe tip.
Today's mainstream atomic force microscopes try to measure atomic force slightly distorting cantilevers, which can be detected by measuring laser light reflected by the distorted cantilever.
↑ But this laser light detection sensor is too bulky to use for multi-probe atomic force microscopes.
Atomic force microscopes with sensitive quartz (= qPlus ) sensors are more compact and known to more precisely distinguish individual atoms (= which can also measure electric current through atoms simultaneously, this or this-p.4-qPlus sensor, this-3rd-paragraph, this-2nd-paragraph ), as shown in sensitive quartz microbalance sensors.
So we should use this current best quartz (= qPlus ) sensor for multi-probe atomic force microscopes instead of the bulky useless laser light sensors ( this-p.7-left-3rd-paragraph, this-p.9-methods ).
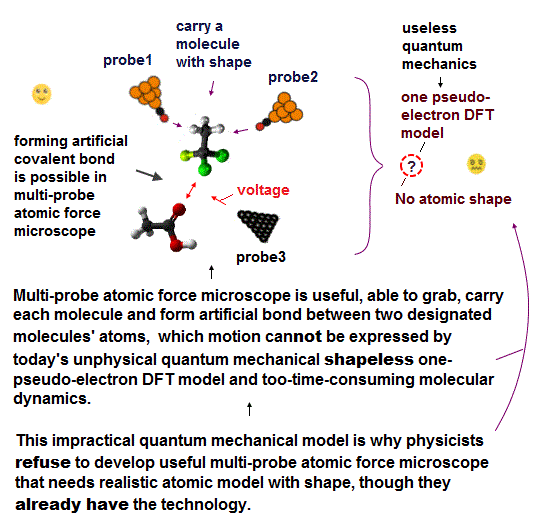
Due to useless Schrodinger equations ( this-1.4 ), today's mainstream physics = fictional quantum mechanics has to treat the whole multiple molecules including a microscope's probe tip's atoms as unreal quasiparticle or one-fake- electron DFT (= Kohn-Sham theory ) model lacking real atomic shape ( this-p.31, this-p.2.1.1 ).
↑ This unreal useless quantum mechanical shapeless atomic model (= which cannot predict anything ) discourages scientists from making useful multi-probe atomic force microscopes that must treat molecules as real objects with shape, though they already have that technology.
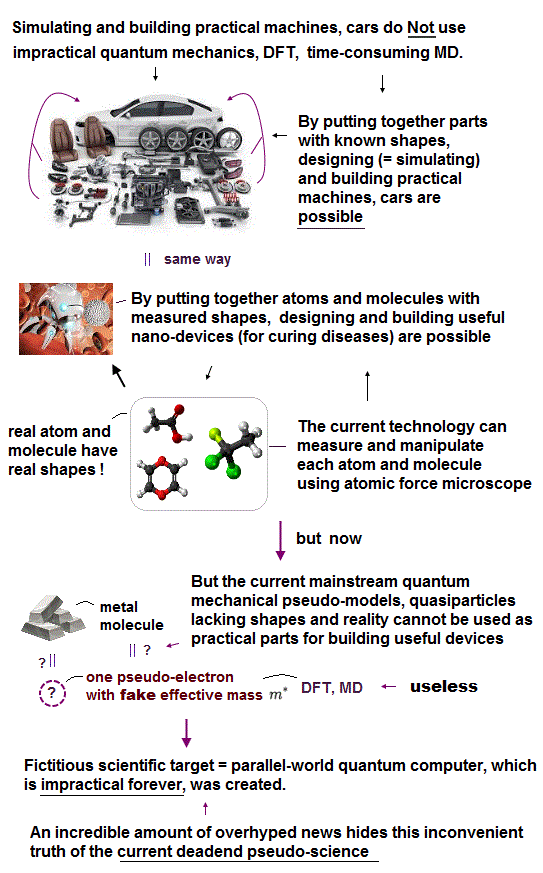
↑ We should discard this current useless quantum mechanical atomic theory, and express observed atoms and molecules as real objects with shape, because we can easily predict reactions and motions of molecules with known shape and properties (= observed by experiments such as multi-probe atomic force microscopes ) like designing and building useful cars (= designing cars' motions does Not use the impractical quantum mechanics ), planes, machines from parts with known shapes.
Due to the current unreal useless quantum mechanical shapeless atomic model hampering developing useful multi-probe atomic force microscopes, today's mainstream atomic force microscopes have only one dull probe tips which can No longer see individual atoms (= which cannot clarify atomic mechanisms nor cure diseases ), which dull probe tips unable to see atoms are often used in biological and medical researches.
So the current atomic force microscopes with dull probe tips allegedly seeing 3-dimensional molecules can only see macroscopic objects, cells, tissues.. without seeing individual atoms ( this-Figure.1, this-p.2-p.3, this-p.6 ).
There are a small number of researches using atomic force microscopes with multiple ( dull ) probe tips, which can only see macroscopic objects or measure electric conductance, cannot see individual atoms.
↑ As a result, the atomic force microscope technology not only stops progressing but also is regressing (= the original atomic force microscopes manipulating individual atoms by sharp probe tips are Not used in most of the current researches ).
We already have the technology of making useful multi-probe atomic force microscopes seeing and manipulating individual 3-dimensional molecules and atoms.
In spite of this fact, strangely, there is No single research paper that even tried to make useful multi-probe atomic force microscopes seeing individual atoms (= there are only a few researches of atomic force microscopes with useless dull probe tips which cannot see atoms ).
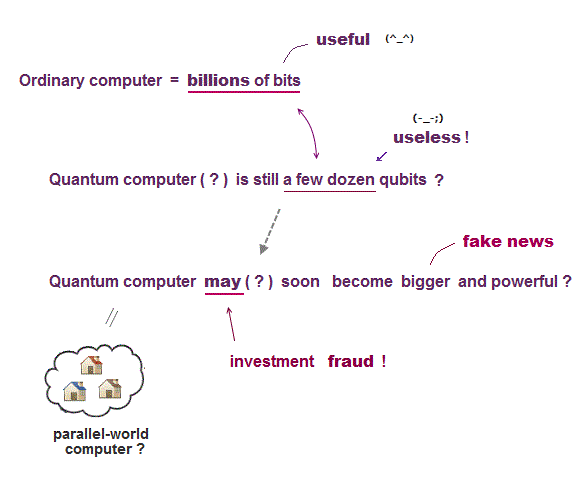
This is in stark contrast to today's mainstream science wasting too much time and money only in overhyped hopeless quantum computers (= scam ) and information, which are impractical forever with No progress.
↑ The academia and journals intentionally block making useful multi-probe atomic force microscopes just to protect their old vested interests and the (useless) quantum mechanics, though we already have the technology of making them.
↑ Actually the possible solutions to the problem of seeing 3-dimensional molecules do Not include multi-probe atomic force microscopes, strangely ( this or this-p.12-3D molecules ).
(Fig.1') Surprisingly, No scientists tried to make multi-probe atomic force microscopes manipulating single molecules, though they already had the technology.
Just to protect the old useless quantum mechanical atomic model such as fictional quasiparticles and one-pseudo-electron DFT, scientists have intentionally avoided making useful multi-probe atomic force microscopes manipulating single atoms or molecules for 40 years.
So the present (fake) multi-probe atomic force microscopes (= AFM ) are used only for measuring electric conductance between multiple probes, or touching objects or cells bigger than single molecules ( this-cell's electrical signal, this-nanowire, this-p.18(or p.7)-p.19 ).
This-p.1-abstract, p.3-3.3 says they just vaguely measured interaction between two (dull) probes instead of measuring single atoms or molecules.
Surprisingly, No scientists tried to make multi-probe atomic force microscopes manipulating single atoms, though they already had the technology.
This or this-p.12-3D molecules section made No mention of making multi-probe atomic force microscopes as solutions to visualizing 3D molecules (← They do Not even try to do it ).
↑ This is in stark contrast to today's physics wasting too much time and money only for the hopeless (overhyped) quantum computers and information that are impractical forever.
(Fig.2) Today's technology can easily prepare sharp probe tips (= cone angle is less than 30o, which is sharp enough to make multiple probe tips ) to which only one atom (= such as a CO molecule, a Cu atom ) is attached, which can precisely manipulate single atoms and build molecular devices.
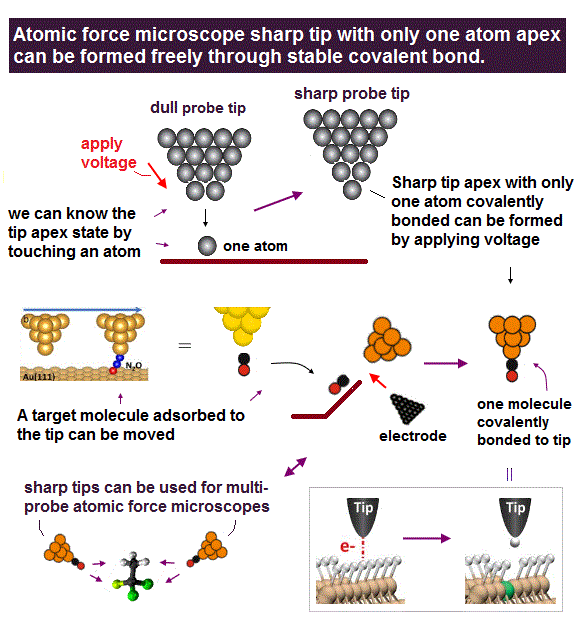
Today's technology can easily make sharp probe tips with cone angle of less than 30 degree, less than 1nm tip radius ( this p.3-Methods, this p.2-last-paragraph, this 8th-paragraph, this-typical spike radius ) whose apex has only one atom such as a CO molecule and a Cu atom.
When the cone angle of the microscope tip is sharp enough or less than 90 degree (= which is possible even without the CO sharper molecular tip ), more than 4 multiple probe tips can be used to touch and investigate a single atom or a molecule at the same time, which means we already have the technology of making useful multi-probe atomic force microscopes.
↑ If we use flat molecules as probe tips, these multiple probes consisting of flat molecular tips (= cone angles less than 180 degree is OK, which is easy to realize now ) can easily avoid colliding with floors.
↑ If we put the target protein onto the slightly elevated floor or sharp molecular floor, we can touch and investigate wider area over the protein by using multiple probes.
We can already attach or adsorb a sharp CO molecule to the apex of a single metallic atom which can be easily prepared by poking probe tips into metallic atoms ( this p.1-last-paragraph~p.2, this p.6-left-method-2nd-paragraph ).
↑ These mono-atomically sharp tips or CO-tips, which are already-existing technologies, are robust enough to manipulate target atoms and molecules ( this p.1-abstract-last, p.2~4 ).
By applying voltage through electrodes, today's atomic force microscopes can form artificial molecular bonds (and bond breakage ), which can be used for making various probe tips with desired atoms or molecules attached ( this p.4-5, this or this-p.17-19, this p.2,p.5-left ).
Whether only one single atom is attached to the apex of a probe tip or not can be easily confirmed by touching the probe tip to a target CO molecule attached to the floor substrate, which is called carbon monoxide front atom identification (= COFI, this p.2, this p.3-right ), as shown in Fig.2-right-lower.
This p.9-methods-3rd-paragraph says -- Single atomic tip possible
"Before we functionalized the tip apex with CO, we prepared metal tips ending in a single Cu atom by repeated
indentations between 300 pm and 1 nm into the Cu surface. Afterwards, we characterized the tip with the
Carbon-Monoxide-Front-Atom-Identification (COFI) method, and repeated the process of tip poking and
COFI characterization until we obtained the COFI portrait of a single-atom metal tip"
Physicists use only big qPlus quartz sensors that are easy to handle, because the quartz sensors are so sensitive that they do not need to make quartz sensors smaller to detect single atoms ( this p.11-13 ).
As shown in quartz crystal microbalance (= QCM ), the smaller and thinner quartz sensors are more sensitive with higher resonator fundamental frequency ( this-p.4-last-paragraph, this-p.2-right-3rd-paragraph, p.3-right-last-paragraph, this-p.2-2nd-paragraph ).
This p.3-left-last-paragraph says -- Sensitive quartz sensor
"Therefore, thinning the
(quartz) oscillator significantly improves the sensitivity of
QCM ( this-p.3-1st-paragraph )"
So we can use the smaller, more sensitive quartz sensors suitable for multiple probe tips of atomic force microscopes.
Atomic force microscope with the quartz tip can touch a single atom and measure small electric current as a scanning tunneling microscope at the same time.
↑ So if we demarcate the floor or substrate (= under target atoms or molecules ) by flowing different patterns of oscillating electric currents in different coordinates, we can easily know the precise position of each probe tip that touches the conducting floor and measures the different electric current's patterns.
(Fig.3) We should use the simpler practical atomic model with shapes which can be easily measured by today's atomic force microscopes as Pauli repulsion, instead of wasting time in useless, time-consuming quantum mechanical model.
Today's atomic force microscopes can easily and precisely measure each atomic and molecular shape as Pauli repulsive force (= unreal quantum mechanical Pauli principle cannot explain this real Pauli repulsive contact force ) or weak van der Waals attraction ( this p.4, this p.4-8, this p.33-45 ).
So we should be able to treat each atom and molecule with known (= measurable ) shape as a real object or part with shape to clarify molecular mechanisms and design molecular devices by using practical multi-probe atomic force microscopes.
The current mainstream old unphysical quantum mechanical models such as Schrödinger equation and density functional theory (= DFT or Kohn-Sham KS theory ) unable to describe each atom as a real atomic object with shape have to rely on extremely time-consuming methods of integrating artificially-chosen (fake) wavefunctions with many freely-adjustable parameters, which is useless, unable to predict anything.
This p.20-last-paragraph says -- Useless quantum mechanics
"KS DFT is Not an ab initio theory (= DFT is just empirical, unable to predict anything, like the useless Schrodinger equations unsolvable for any multi-electron atoms ), because it does
not approach the exact solution"
↑ To protect this current too-time-consuming impractical quantum mechanical models lacking real atomic shape such as DFT, MD ( this-p.9-12 ), academia intentionally hampers developing the useful multi-probe atomic force microscopes that will make the useless quantum mechanics unnecessary and obsolete.
(Fig.3') Today's atomic force microscope with only one probe tip cannot see 3-dimensional (= 3D ) molecule, so useless.
Atomic force microscopes manipulating single atoms have had only one useless probe tip for 40 years with No progress.
This current atomic force microscope with only one probe can not see 3-dimensional (= 3D ) molecules ( this or this-lower-challenges, this or this-p.12-3D molecules ), because the target molecule pushed by one probe is easily moved or rotated unstably, which cannot get clear atomic structures.
Due to the useless one-probe atomic force microscopes, today's medical researches use only (useless) atomic force microscopes with dull probe tips that can vaguely see only cells and objects far bigger than single atoms ( this-figure1 ) = cannot see individual atoms. = Our nano-technology is regressing.
Only atomic force microscopes with multiple single-atomic probe tips can see 3-dimensional (= 3D ) atomic structures by one probe fixing the target molecule stably while another probe touching and seeing atoms.
↑ A whole molecule is often rotated around some rotation axis when pushed by a probe tip.
The current unreal quantum mechanics treating all molecules (= including a microscope's probe tip's atoms ) as one-fake-electron (= DFT, quasiparticle ) model lacking atomic shape can Not express target molecules moved or rotated by microscopes' probe tips.
In order to describe molecules rotated by probe tips, we have to define a real shape of a molecule and the rotation axis of the whole rotatable molecule, which is impossible in the unreal quantum mechanical shapeless atomic model.
As shown in the current only molecular motion simulation method = impractical molecular dynamics, quantum mechanics can only give fictitious potential energy (= Not real shape ) to each atom, which unreal shapeless quantum mechanism molecular model cannot even predict how a molecule is moved or rotated when one atom of the molecule is pushed by the microscope's tip.
↑ So today's (useless) atomic force microscope with only one probe tip can only see flat static molecules such as benzenes or only upper parts of stably-fixed molecules ( this-p.4-p.5, this-p.2 ), which cannot be applied to practical use.
To protect this useless old quantum mechanical atomic model, academia has intentionally hampered making useful multi-probe atomic force microscopes (= which have to treat molecules as real rotatable objects with shapes without unreal quantum mechanics ), though we already have that technology (using sensitive quartz sensor, this-p.7-9 ).
We should use actual atomic shapes obtained by experiments (= multi-probe atomic force microscopes ) to develop nano-technology, which is hampered by the unreal quantum mechanical shapeless atomic wavefunctions.
(Fig.3'') Quantum mechanics is useless, too time-consuming.
Due to the impractical Schrodinger equations, Quantum mechanics has to artificially choose one-pseudo-electron DFT wavefunctions (= ψ ) expressing the whole molecules and probes' atoms (= these shapeless wavefunctions hamper science ), and integrate them to get fake total energy.
↑ This present mainstream quantum mechanical one-fake-electron DFT (= Kohn-Sham theory ) approximation, which must artificially choose fake exchange correlation-potential energy (= functionals ), is empirical or fake ab-initio, which cannot predict any molecular energy ( this-p.2-last, this-p.18, this-p.1-introduction ).
Quantum mechanics can only express the whole multiple molecules and materials as one-fake-electron DFT wavefunctions consisting of multiple coefficient free parameters (= c ) and artificially-chosen basis set functions (= different DFT wavefunctions must be chosen in different atomic positions ), which cannot predict anything.
And they differentiate the energy equation (= integral of chosen one-fake-electron wavefunctions with DFT energy equation including chosen fake exchange energy functional ) with respect to coefficient parameters (= c ) of the chosen wavefunction or basis sets ( this-p.5 ) to find coefficients that give the lowest (fake) atomic energy within the chosen wavafunction in variational method ( this-p.5-5 ).
See this-(1)~(24), this-p.59(or p.49),p.93, this-p.32-2.48,
They have to repeat this extremely time-consuming meaningless quantum mechanical methods of integrating and differentiating the DFT energy equations until the coefficient parameters luckily converge to some values, which old inefficient method is called self-consistent-field (= SCF, this-p.9, this-p.2-p.8, this-p.6-p.18, this-p.3 ) iteration hampering science.
The present extremely-time-consuming impractical quantum mechanical SCF (= self-consistent-field ) iterative calculations repeatedly updating the chosen (fake) wavefunctions' coefficient free parameters often fail to get the converged parameters of the chosen fake wavefunctions (= so No atomic energy E is obtained by this impractical quantum mechanics, this-middle ).
This-lower SCF convergence says -- Impractical quantum mechanics
"Conventional electronic-structure-theory-based methods like DFT rely on a self-consistent-field (SCF) process, where an initial guess for the electron density is generated and then iteratively refined through repeated computation of electron–electron repulsion. This process frequently exhibits chaotic behavior, and SCF convergence can become extremely time-consuming or even impossible."
For the unphysical quantum mechanical shapeless atomic model to explain the interaction between the molecule and atomic force microscope's probe tip, they have to repeat this extremely time-consuming SCF energy calculations by choosing different fake wavefunctions (= and repeatedly updating their coefficients parameters until they converge ) in many different positions (= A, B, C, D ) of the atoms of the molecule and the tip.
↑ Comparing energies of artificially-chosen one-fake-electron DFT wavefunctions in different atomic positions and finding the lowest-energy atomic-position state is called geometry optimization which is impractical, too time-consuming, unable to predict real molecular energy.
↑ This impractically time-consuming repeated SCF energy calculations by choosing many different one-fake-electron-DFT wavefunctions (+ repeatedly updating parameters ) in many different atomic or tip's positions to compare (fake) energies E in different atomic positions and find the lowest-energy atomic position state is called geometry optimization ( this-last-calculation method ) or relaxation ( this-p.27-53, this-p.3, this-p.18, this-p.6-2nd-paragraph ).
This-p.1-1 intoduction~p.2 says -- Impractical geometry optimization
"Geometry optimization is a process to find the atomic coordinates that minimize the energy
of a target molecule. It is an important basis task in quantum chemical calculations because the optimized geometries are used to analyze the molecular characteristics and structures.
In geometry optimization, a stationary point on a potential energy surface (PES) is explored
by iteratively calculating the energy and gradients of a molecule while changing its atomic
coordinates step by step"
↑ This quantum mechanical time-consuming geometry optimization is said to be able to find the most stable atomic positions with the lowest energy E (= after comparing energies E in different atomic positions ), but actually their artificially-choosing one-fake-electron DFT wavefunctions and exchange pseudo-potential energy can Not predict any atomic positions.
This-3rd-paragraph says
"the geometry optimizer proceeds to update the atomic positions and rerun ( time-consuming ) SCF (= which often fail, this-p.22, this-p.31-2nd-paragraph )."
It is far better to use experimental values (= each molecular shape and properties ) from the beginning than to waste too much time in these meaningless time-consuming quantum mechanical calculations.
In this impractical time-consuming quantum mechanical geometry optimization, the final seemingly-stable atomic positions of a molecule is just one of many local energy minima (= Not the lowest-energy nor most-stable atomic positions ) instead of a global energy minimum, so initially-chosen one-fake-electron wavefunctions or initial-guess of atomic positions influence the final positions ( this-p.3-last-paragraph ), which cannot predict real atomic positions.
This site on geometry optimization ↓
1st-paragraph says -- Incorrect energy
"geometry optimization, molecular optimization or relaxation, is the process by which the geometry of a molecule is adjusted to find a structure with the lowest possible energy. This optimized structure typically corresponds to a stable configuration, either a local minimum (= Not the global energy minimum state, so No correct solution is obtained )"
Point to remember says -- Useless geometry optimization
"Geometry optimization seeks the nearest local energy minimum or transition state. This means that the resulting structure is dependent on the starting geometry. Different initial geometries may lead to different optimized structures.
The chosen quantum mechanical method and basis set play significant roles in the accuracy and reliability of the optimized geometry.
While the goal is to find the global minimum (the absolute lowest energy structure), in practice, geometrical optimization often finds local minima"
↑ So the quantum mechanical geometry optimization often gets wrong energy solutions = local-energy minima, which depends on artificially-chosen wavefunctions, which cannot predict any real molecular energy or atomic positions.
This or this-p.19-last says -- No solution
"None of these (= quantum geometry optimization ) are guaranteed to find the global
minimum (= the lowest-energy atomic position state or solution cannot be obtained ) !"
This-p.4-right-2nd-paragraph says -- Too many impractical SCF
"the calculation of forces on one iteration of
the geometry optimization requires 6N +1 separate SCF runs
where N is the number of atoms" ← Just calculating force on each atom requires 6 × time-consuming repeated SCF energy calculations of artificially-chosen wavefunctions, impractical.
↑ One SCF calculation takes about 2300 seconds (= this-last-paragraph, = 40 minutes ).
This-p.6-4th-paragraph says -- Too time-consuming
"Calculations for determining only one torsional angle by comparing energies of different angles or different atomic positions (= each 10o step ) by DFT SCF and geometry optimization took 34679.9 seconds (= 9.3 hours )"
This-p.3-upper says -- Impractical DFT
"in the determination of the tip–sample
forces,... This can be done with standard DFT codes but it's time consuming"
(Fig.T) practical atomic model with shape vs. useless too-time consuming quantum mechanical shapeless wavefunction.
The current impractical quantum mechanics just choosing fake wavefunctions and pseudo-potentials is too time-consuming, cannot predict any atomic (or protein ) energy or structures ( this-p.35, this-p.1-right ).
This-p.1-left-2nd-paragraph says -- Impractical DFT
"A quantum mechanical method to analyze
molecular vibrations is density functional theory (DFT).
Because of the large computational cost, DFT calculations
cannot be applied to entire proteins"
This-p.2-introduction-1st-paragraph says -- Impractical quantum
"Molecular energy calculation is crucial for conformation analysis and structure-based drug
design. However, accurate calculation achieved by high-level quantum mechanical
calculations1
can be computationally demanding even for small molecules,
while the
application of more computational efficient molecular mechanical methods
is limited by
the accuracy of force fields"
Actually the present overhyped AI, Alphafold, which must rely on experimentally-obtained training datasets, cannot predict unknown proteins' structures nor motion due to lack of experimental data ( this or this-lower-Limitations and challenges, this-lower-limitation ).
Overhyped quantum computers are also impractical forever.
Only multi-probe atomic force microscopes can obtain the exact molecular or protein atomic structures, which are hampered by the unreal quantum mechanical shapeless atomic model.
We can easily predict how a molecule or a thing with known shape is moved or rotated when pushed by a probe, which is impossible in the current unreal quantum mechanical shapeless atomic model.
Quantum mechanics can only give artificially-created potential energy called force fields to its shapeless atoms, which takes too much time to simulate actual molecular or protein motion in today's only molecular-motion simulating method = impractical molecular dynamics (= MD, this-3rd~5th-paragraphs, this-p.1-right-1st-paragraph ).
↑ This current impractical MD takes more than days to simulate just 1-microsecond molecular motion, which cannot simulate or explain actual molecular motion of seconds ~ hours observed by atomic force microscopes (= AFM, this-p.2-Figure 1a ).
We have to estimate how we push each molecule (= with known shape ) to get its desired motion for practical application of molecules, which is impossible in today's unreal quantum mechanical shapeless atomic model where we have to blindly try infinite different patterns of pushing the shapeless molecule to get some desired molecular motion, which takes infinite time in repeating the impractical MD molecular-motion simulations.
For example, the geometry optimization took extremely much time = a week for this impractical quantum mechanical SCF to calculate and compare various energies in different atomic positions (= after choosing different one-fake-electron DFT wavefunctions ) and find the most stable energy state of some small molecule observed by the atomic force microscope (= AFM ) with only one tip. ↓
This-p.4-5th-paragraph & p.5-2nd-paragraph-last say
"The
interaction energies of the tip and the sample molecule (without substrate) were
calculated for various lateral positions with a lateral spacing of 0.1 Å and for tip heights
ranging from d = 3.95 Å to d = 3.35 Å with a vertical spacing of 0.025 Å. To obtain the
frequency shifts (= Δf, p.3-top ), the interaction energies were numerically differentiated twice with
respect to d"
"The calculation of the Δf(x) line profiles (= frequency f shift obtained by differentiating the artificial molecular energies calculated by DFT in various different tip positions ) took about a week computation time on a large computer facility. ← too time-consuming, impractical ( this-p.3-1st-paragraph )."
This-p.2-right-1st-paragraph says -- Calculations tool a year !
"to
explore the topology of the PES (= potential energy surface ) of docosahexaenoic acid (DHA)
With DFT methods, this analysis would require more than a
year of computation time" ← too time-consuming, useless for any applied science, medicine.
Today's only method of simulating motions of the (unreal) quantum mechanical shapeless atoms is the impractically-time-consuming molecular dynamics ( this-p.5-102-sentence says just 50ps molecular motion was simulated, which is impractical ).
We should replace this impractical time-consuming quantum mechanical wavefunction by the much simpler atomic model with experimentally-measured shape for the practical multi-probe atomic force microscopes to clarify complicated proteins at the atomic level and cure diseases.
(Fig.4) The use of many-probe, multi-probe atomic force microscopes can efficiently, speedily generate artificial chemical reactions and desired molecular bonds.
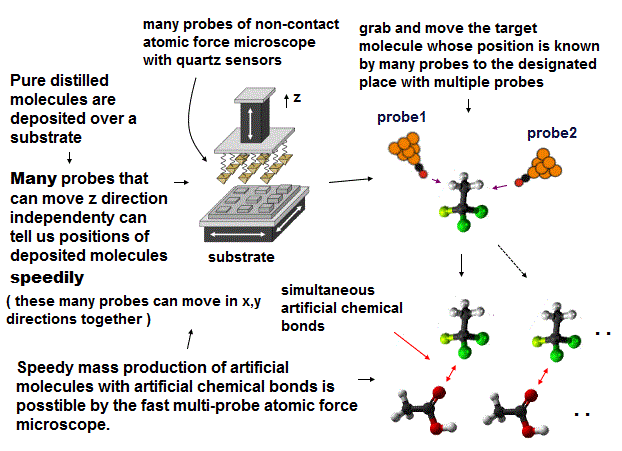
Unlike the useless one probe, atomic force microscopes with multiple probes can observe, analyze, manipulate any forms of molecules and proteins (= in case of a large protein, breaking the protein little by little using artificial molecular bond dissociation through applying voltage enables us to know the precise outer and inner protein structure by the multi-probe atomic force microscope, this or this-p.21 ).
Of course, the practical multi-probe atomic force microscope can design and build useful molecular devices (= used for artificial efficient photosynthesis based on artificial catalysts and curing cancers ) by manipulating single atoms, using realistic atomic model with shape (= instead of the unphysical impractical quantum mechanical shapeless atomic model ).
Efficient speedy chemical reactions forming artificial covalent bonds between the designated atoms of molecules, which are impossible otherwise, are possible using multi-probe atomic force microscopes.
In the process of the speedy artificial chemical (= molecular bond ) reactions, first we deposit distilled pure molecules over a substrate (= by vacuum deposition or something ).
If we put some different marks or narrow lines (< 10nm ) with slightly different heights (or different lengths, widths ) or consisting of different atoms on different positions of the substrate, we can know the positions of each probe (and deposited molecules ) immediately when tips touch the substrate surface.
Next, we can know the positions of target molecules deposited over the substrate by using non-contact (= high-resolution quartz sensor ) atomic force microscope with many probe tips (= each probe can move in z direction independently to detect target molecules with different heights automatically at very high speed. All these many probes are moved together in x,y directions ).
↑ A probe tip can be moved (= displaced ) up to the distance of 0.1% of the piezo electric material (= actuator ), so moving atoms 1μm needs 1mm piezo electric material ( this p.3-left, this 4~6th-paragraphs ).
↑ Moving each probe of many probes in x-y directions independently needs much space of the piezo material (= 1mm × 1mm ) in each probe, while moving each probe only in z direction independently does not need so much space (= just tiny 1μm × 1μm space in x,y directions is OK ), which can increase the dense of probes (= many compact probe tips ) and speed up the atomic force microscope finding all target molecules deposited over the substrate.
After knowing the positions of target molecules on the substrate by many probes, we can move those molecules by an atomic force microscope with multiple probes (= manipulating single molecules freely in x,y,z directions ) from the substrate to some designated places where many artificial covalent bonds can be formed simultaneously by applying voltages between two molecules placed on designated positions.
As a result, the fast, practical mass production of artificial molecules with desired covalent bonds (= which are impossible in other ways ) is possible in the atomic force microscope with multiple probe tips manipulating single molecules.
(Fig.5) Speedy efficient chemical bond formation by nano-plates built by multi-probe atomic force microscopes
As shown in the upper figure, if we build nano-plates designed to stick to some molecules by multi-probe atomic force microscopes, we can easily and efficiently cause artificial chemical reactions or bonds between some specific molecules by controlling two nano-plates designed to stick some molecules on designated positions and flowing electricity.
↑ These artificial nano-plates or enzymes built by multi-probe atomic force microscopes can be repeatedly used, and kept clean for a long time by using cathodic protection.
By using practical multi-probe atomic force microscopes, we can clarify detailed atomic mechanisms of proteins in various diseases, and design some effective drugs or molecules sticking only to some cancer cells, HIV-infected cells, and auto-immune-antibody-producing cells, and remove them (= after these artificial molecules stick to target sick cells, only these sick cells are prevented from dividing, and eventually die, which can cure the current incurable diseases ).

Feel free to link to this site.