Home page
Quantum biology is useless
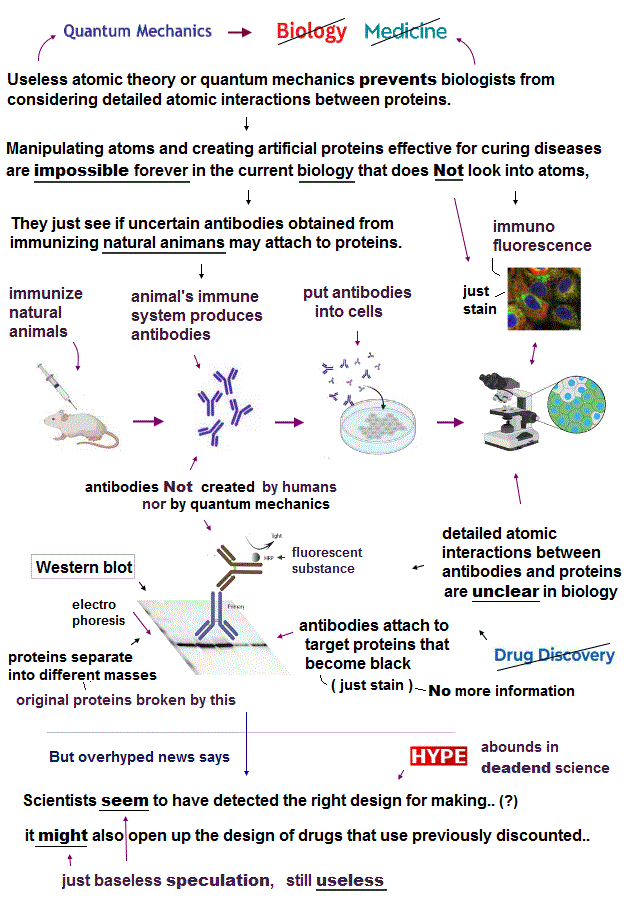
(Fig.1) Useless quantum mechanics hampers medicine from clarifying atomic mechanisms of diseases.

The present biological, medical researches use only macroscopic tools obtained from natural organisms such as PCR enzymes, DNAs, antibodies, cells, mice.. that do Not see atoms nor use ( useless) quantum mechanics ( this-4.Biologists' views, this-p.30-10-conclusion ).
This-p.6-fifth-paragraph says -- No quantum biology
"for the most part biologists can ignore quantum mechanics; the practice of biology was not revolutionised by the discovery of
quantum mechanics ( this-Two arguments against the quantum brain )"
The fact that today's medicine cannot see nor clarify atomic mechanisms is why many drug clinical trials failed, and cancers, Alzheimer are still incurable.
In biological research, target DNAs or genes are amplified by PCR (= polymerase change reaction ) enzymes (= isolated from bacteria Not made from quantum mechanics ), and inserted into virus plasmid DNA vectors where several different proteins' genes are artificially connected (= cloning ), which produces the genetically-modified proteins ( this-p.2~ ).
These proteins are vaguely seen, sticking to antibodies ( this-p.10 ), which are created by immunizing animals with the target protein's antigens, attached to fluorescent molecules and proteins such as GFP (= green fluorescent proteins obtained from jellyfish Not made from quantum mechanics ) through fluorescent optical microscopes that can Not see atoms ( this-p.6~ ).
↑ These biological methods of vaguely seeing fluorescent antibodies (= Not seeing detailed atomic structures ) sticking to target proteins include immunofluorescence ( this = this-p.13 ), Western blotting ( this-p.3, this = this-p.6 ), immunoassay, immunoblotting ( this = this-p.12 ), ELISA ( this-p.3 ), flow cytometry = FACS ( this = this-p.19 ), immunoprecipitation ( this = this-p.21 ), and immunohistochemistry ( this = this-p.6 )
Actually today's biological or medical researches do Not use any quantum mechanical methods such as Schrödinger equation and density functional theory (= DFT ) that are Not mentioned in these biological papers (= this-p.2-Materials and methods, this-p.8~9-Materials and methods used only macroscopic biological tools )
Biological and medical researches often use fluorescent optical microscopes with too bad resolution (= 200nm, this-3rd-paragraph ) to see individual atoms (= each atomic size is only 0.1nm ).
So medical researches can Not see nor clarify atomic mechanisms of diseases, hence curing difficult diseases is impossible.
In RESI (= resolution enhancement sequential imaging ), target proteins are attached to some tag molecules with fluorescent molecules or proteins.
↑ This RESI can distinguish only different fluorescent molecules (= can see only vague colors, Not see individual atoms, this-p.2~3 ) with better resolution, but this method can Not see target proteins themselves, so useless.
This-7~9th-paragraphs say -- Not see atoms
"The size of the tag is also a limiting factor because it is the position of the fluorescent part of a tag that is observed, rather than the molecule of interest that the tag is connected to."
The best microscopes used in today's biological and medical researches are cryo-electron microscopes (and X-ray crystallography ) whose resolution is too bad (> 0.4nm ) to see individual atoms of 0.1 nm (= 1Å, this-p.11-conclusion, this-lower-Challenges and limitation ).
As a result, today's medical, biological researches (and AI, Alphafold needing experimentally-obtained protein structures ) can Not see nor clarify atomic mechanisms, so can Not cure the current intractable diseases such as cancers, Alzheimer, HIV, autoimmune...
Only atomic force microscopes (= AFM ) can see and manipulate individual atoms, but can see only flat molecules such as benzenes ( this or this-lower-challenges, this-p.12-3D ), because an atomic force microscope has had only one probe tip for 40 years with No progress.
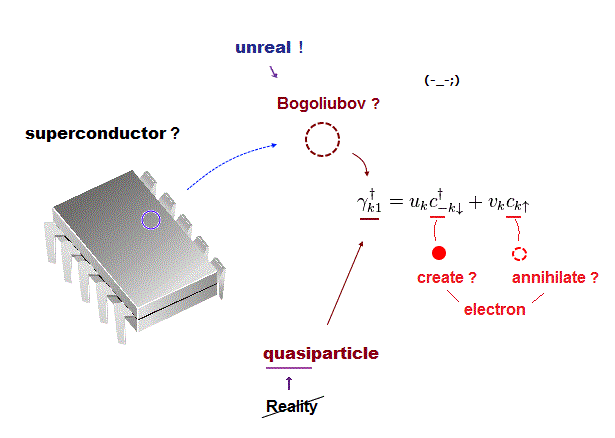
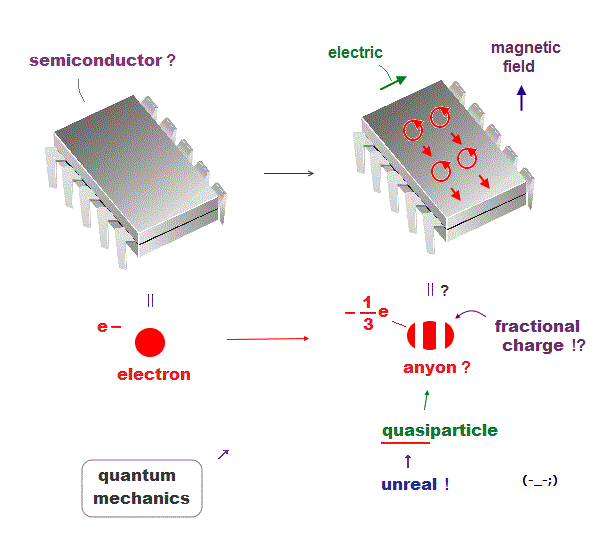
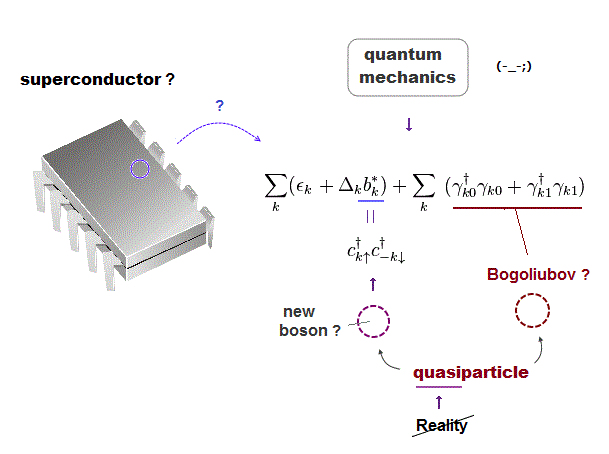
Today's only atomic theory = very old fictional quantum mechanics can describe molecules or proteins only as unreal quasiparticle models or one pseudo- electron DFT (= density functional theory ) model lacking real atomic shape.
↑ These quantum mechanical Schrodinger equations and DFT are not only unable to predict any physical values but also hampering nanotechnology due to their unrealistic shapeless atomic model.
↑ Useful atomic force microscopes need multiple probe tips manipulating any molecules and proteins freely by realistic atomic model with shape (= obtained from experiments rather than theory ) that clearly contradicts today's unrealistic quantum mechanical shapeless wavefunction.
↑ So the present academia intentionally prevents making useful multi-probe atomic force microscopes just to protect the old impractical quantum mechanical model.
(Fig.2) Biological tools do Not rely on (useless) quantum mechanics nor consider microscopic atomic interaction.
Medical and biological researches use only macroscopic procedures and tools originating from natural organisms such as bacteria and virus irrelevant to (useless) quantum mechanics.
The so-called quantum biology is just a useless pseudo-science irrelevant to real biology like impractical quantum computers.
↑ This misleading quantum mechanical biotechnology often mentions magnetic sensors such as MRI and NV-center in diamond that can be explained by classical mechanics, and these quantum sensors (= just vaguely detecting external magnetic field ) cannot clarify atomic mechanisms of biological reactions.
Actually No quantum mechanical methods = Schrödinger equation or DFT (= which are useless, cannot predict anything ) are used in today's biotechnology researches ( this-p.6~8, this-p.10~11 ).
(Fig.P) PCR uses DNA polymerase enzymes originating from natural bacteria, Not from (useless) quantum mechanics.
PCR or polymerase chain reaction is one of most popular biological methods for increasing and amplifying target DNA genes after preparing short DNA primer genes (= attach to both edges of target-protein genes ) by using enzymes called polymerase originating from natural bacteria (or natural polymerase whose gene is slightly modified using other natural enzymes ).
↑ No quantum mechanical methods are involved in discovering or developing PCR enzymes.
Even the misleading research on unreal negative-kinetic-energy quantum tunneling ( this-p.5-conclusion) says -- Unclear quantum effects
"However, it's important to acknowledge the
limitations of the model in drawing definitive conclusions. The
complex oscillations and behaviors observed during PCR, combined
with the temperature-dependent nature of quantum effects, suggest
intricate interactions that require further investigation. The study
underscores the need for more comprehensive experimental
validations and refined models"
RT-PCR or reverse transcription polymerase chain reaction is the method of detecting RNA genes such as COVID-19 viruses by changing the target RNA into complementary DNA (= cDNA ) and amplifying those cDNA using ordinary PCR.
The enzymes called reverse transcriptase used in this RT-PCR were derived from natural retroviruses (or their slightly-gene-modified enzymes), Not from being designed or created by (useless) quantum mechanical calculation.
Real-time PCR is the method of quantifying the target DNA genes by using ordinary PCR and fluorescent molecules. ← All of these biological techniques are developed based on actual experience and experimental observations, and No quantum mechanical prediction is involved.
(Fig.P) DNA sequencing also uses DNA polymerase from bacteria.
Sequencing is a technique used for reading and determining the order of nucleotides (= adenine-A, guanine-G, cytosine-C, thymine-T ) in the target DNA or RNA.
DNA sequencing uses the ordinary DNA polymerase and "stop-nucleotides labeled with different-color fluorescent molecules (= A, G, C, T nucleotides are labeled in different colors ), measures the random lengths of randomly-amplified DNA fragments by electrophoresis, identifies what nucleotide is at the edge of each DNA fragment by measuring colors, which eventually could determine the whole target DNA's sequence or order of nucleotides.
No quantum mechanical calculation is involved in this technique.
RNA-sequencing is also for determining RNA nucleotides by changing the target RNA into cDNA by reverse transcriptase and using the method similar to DNA sequencing.
The examples of researches using this sequencing are this = this p.11
DNA microarray is a technique to see if target DNA can bind to some complementary nucleic acid sequences based on experimentally-known complementarity nucleotide pairs ( A-T, G-C ), not on quantum mechanics.
Small interfering RNA (= siRNA ) or microRNA is a short nucleotide RNA molecule used in silencing or suppressing the expression of the target genes by binding to target nucleotides through the well-known RNA complementarity, Not by quantum mechanical calculation.
This siRNA, microRNA or gene silencing utilizes the mechanisms inherent in natural organisms, whose silencing mechanism was Not created or designed by human technology from scratch or useless quantum mechanical calculations.
Short-hairpin RNA or shRNA is also one of these siRNAs that utilizes natural organisms' gene silencing mechanism and enzymes.
To roughly know the amount of DNAs and RNAs, the absorbance of lights with some specific wavelengths are used.
Rough determination and quantification of total protein's concentration also uses the similar measurement of light absorption called Bicinchoninic acid assay.
↑ All these methods are unable to determine precise structure or identify the target protein, and these techniques are based on empirical observations, Not on (useless) quantum mechanical calculations.
(Fig.S) SDS-PAGE just roughly separates proteins by size, No atomic interaction nor (useless) quantum mechanics is considered.
SDS-PAGE is an electrophoresis method that separates proteins by masses = bigger proteins move slower than smaller proteins, which is common sense by classical mechanics.
This SDS-PAGE or electrophoresis cannot know the protein's detailed structure or atomic interaction. ← inapplicable to developing drugs
(Fig.L) Liquid chromatography separating mixed proteins based on affinity to some molecules (= such as antibodies and water ) is irrelevant to quantum mechanics.
Protein chromatography separates proteins by their different sizes, different affinities with different ligands or antibodies or light scattering based on experimental observations and general knowledge.
Each molecule and protein always sticks to other molecules and proteins, so complete separation or avoiding contamination is impossible.
No quantum mechanical calculations are involved.
(Fig.M) Mass spectrometry (= MS ) separating proteins based on masses is irrelevant to quantum mechanics.
Mass spectrometry (= MS ) is the method of determining proteins' masses by ionizing target proteins and fly them under electric field where heavier proteins move slower (or harder to accelerate ) than lighter proteins. which is obvious in classical mechanics. Quantum mechanics is unnecessary.
This MS just estimating masses cannot know the detailed protein atomic interactions (ex. this = this p.15-right, this = this p.7-9 ).
LC-MS/MS mixing the above liquid chromatography and mass spectrometry is often used to separate and vaguely identify molecules and peptides of mixed proteins.
Proteosome assay roughly measures the activity of natural proteasome enzymes by using substrate and fluorescence ( this p.3 ), based on empirical observation, Not on (useless) quantum mechanical calculations.
EMSA (= electrophoretic mobility shift assay ) is used to see if DNA roughly binds to proteins (= estimating DNA-protein interaction ) by using electrophoresis where proteins bound to DNA become heavier and move slower, which is common sense.
This method cannot know the detailed atomc interaction mechanism between DNA and proteins, nor use (useless) quantum mechanics.
ATP assay is a method of roughly estimating the amount of natural molecules called adenosine triphosphate (= ATP ) by using luciferase enzymes originating from natural organisms and measuring emitted light.
(Fig.S) CRISPR-Cas9 for gene editing was obtained from natural bacterial immune system, Not from (useless) quantum mechanics.
CRISPR-Cas9 system used in gene editing was obtained from natural bacterial immune systems, Not from being artificially designed or created by humans or useless quantum mechanical calculations.
Synthesis of mRNA (= messenger RNA ) by DNA → RNA transcription in vitro uses enzymes called RNA polymerase originating from natural organisms such as bacteriophages (e. T7 bacteriophage or T7 phage ) that are natural viruses infecting bacteria or Escherichia coli ( this introduction ) combined with natural nucleotides such as ATP, CTP, GTP.. ( this p.5, this = this p.9-right ).
These enzymes were Not designed or created by humans nor by (useless) quantum mechanical calculation.
(Fig.M) Today's medical research cannot utilize atomic interaction due to useless quantum mechanical atomic theory, so clarifying real atomic mechanisms of intractable diseases is impossible.
The 2nd and last paragraphs of this hyped news say
"Researchers.. have developed the first methods to identify and characterize all the various irregular forms of amyloid beta"
"Together, these results are really encouraging for our quest to better understand Alzheimer's disease" ← just "encouraging", still Alzheimer is incurable.
↑ This research paper ↓
p.1-abstract-last says "Herein, we used immunoprecipitation to examine the binding affinity of four antibodies against 18 epimeric and/or isomeric Aβ peptides (= amyloid beta seen in Alzheimer ).. Tandem mass spectrometry was used as a detection method" ← Not seeing atoms.
p.6-7 Methods did Not use any (useless) quantum mechanical methods such as Schrodinger equations and density functional theory (= DFT ).
The 2nd-last and last paragraphs of this hyped news say
"The results of this work also show that ERK5 inhibitors would (= just speculation ) potentiate the anticancer activity of NK cells."
"In the future (= still unrealized now ), the researchers hope to find new collaborations to conduct clinical trials, with the aim of testing ERK5 inhibitors in cancer patients."
↑ This research paper ↓
p.11-right-DNA constructs used plasmid DNA vectors based on natural viruses, No quantum mechanics.
p.13-left used bacterial CRISPR, immunoblotting, immunoprecipitation based on antibodies obtained by immunizing natural animals. ← All these biological tools originated from natural organisms Not from (impractical) quantum mechanical technology such as Schrodinger equation and one-pseudo-electron DFT model.
p.13-right-1st, 3rd, 5-6th paragraphs also used flow cytometry, immunoprecipitation, immunoblotting based on natural animals' antibodies.
p.15 used SDS-PAGE electrophoresis, liquid-chromatography, mass-spectrometry, RNA-sequencing, immunophenotyping (= with antibodies ), .. all of which are macroscopic biological experiments without looking into detailed atomic interaction nor (useless) quantum mechanical calculation.
The 1st, 10th paragraphs of this hyped news say
"the current treatments fall far short of being effective at regaining memory." ← Alzheimer diseases is still incurable despite longtime researches.
"KIBRA (= protein ) restored synaptic function and memory in mice, despite not fixing the problem of toxic tau protein accumulation (= cannot fix Alzheimer ).. Our work supports the possibility (= just speculation, still useless ) that KIBRA could be used as a therapy to improve memory"
↑ This research paper ↓
p.16-left used immunolabeling, Western blotting by antibodies (= obtained from immunizing natural animals ), GFP (= green fluorescent protein obtained from jellyfish ). ← Not seeing atoms nor using quantum mechanics
p.16-right used immunoblot, immunostaining based on antibodies.
p.17-left-1st,2nd-paragraphs also used Western blot based on antibodies.
↑ No quantum mechanical methods such as Schrodinger equation or DFT were used.
And No detailed atomic interaction was considered (= atomic mechanism remains unclear ) even in this recent Alzheimer research, which is why this disease is still incurable.
This recent research on brain diseases also did Not use the useless quantum mechanics, DFT, MD, Alphafold nor clarify atomic mechanism. Only macroscopic biological tools were used ( this-p.14~17 materials and methods ).
(Fig.M) Today's overhyped biological research just relies on old-fashioned macroscopic experiments based on natural-organism-based viral DNA vectors, cells, bacterial proteins without considering atomic interaction, which cannot develop effective drugs..

The 4th, 9th, last paragraphs of this hyped news say
" the proteins MinD and MinE—known collectively as MinDE—interact with each other along the cell membrane to produce wave-like patterns, which aid in the movement of molecules within the cell."
"By engineering interactions between the MinDE proteins and proteins of interest, the researchers created highly specified patterns to organize molecules within mammalian cell"
"The Coyle Lab plans (= still unrealized, No practical application ) to continue exploring the tool's applications"
↑ This research paper ↓
p.2-Main text-4th-paragraph says -- Biology Not seeing atoms
"We used lentivirus to express mCherry-MinD and MinE-GFP in multiple different mammalian cell lines and
imaged their spatiotemporal distribution by timelapse fluorescence microscopy (= cannot see atoms )"
↑ This research just joined bacterial oscillating proteins MinD and MinE genes to jellyfish's fluorescent GFP gene and sea anemone's fluorescent mCherry gene inside lentiviral plasmid vector which infected mammalian cells to express these proteins (= called transduction ).
The expression of these bacterial MinDE proteins joined to fluorescent proteins in mammalian cells was observed by microscopes. That's all. No atomic interaction was considered nor clarified.
↑ All these biological tools for gene editing in lentiviral DNA vectors originated from natural organisms, Not from humans' design.
No quantum mechanical methods (= Schrodinger equation, DFT.. ) were used in this biological research.
The 2nd, 3rd, 7th, 13th paragraphs of this hyped news (7/1/2025) say
"This approach could (= just speculation ) help scientists study how molecules associate inside cells," ← No detailed practical use is mentioned.
"use natural proteins produced by a cell (= proteins Not designed by humans nor by useless quantum mechanics ) as tiny sensors to report on their environment and interactions, "
"By triggering the flavin's magnetic properties with light, they could use ESR to study protein structures directly inside cells"
"a small engineered flavoprotein called iLOV, which can be genetically fused to other proteins to make them visible with ESR."
↑ So this research just measured ESR (= magnetization caused by electron's orbit, Not by the unrealistic electron spin ) in the natural flavoprotein fused with other proteins in cells by macroscopic biological methods without using the (useless) quantum mechanics.
↑ This-p.9-11-methods used only macroscopic biological tools, did Not use any quantum mechanical methods.
The 3rd, 6th-last, 2nd-last paragraphs of this hyped news (7/2/2025)
"By tracking the movement of RNA molecules inside the nucleolus using advanced imaging and genomics techniques, the new method allows researchers to watch these processes as they unfold without destroying the cell"
"to disrupt different steps in the ribosome assembly line and use a system called a DNA plasmid (= originating from natural viruses ) to induce living cells to create brand new, human-designed nucleoli." ← No. The present useless quantum mechanics prevents humans from creating really-artificial nucleoli
"Now that they have these tools, ..his group are looking at what happens in diseases like cancer,... they hope (= still unrealized ) to find vulnerabilities in the production process which could be targets for therapeutics."
↑ This research paper ↓
p.11~ Methods -- Not seeing atoms
used only macroscopic biological tools such as cells, rDNA (viral) plasmid construction encoding natural nucleolus RNAs (= so No really-artificial nucleolus was created in this research ), immunofluorescent (= IF ) by animal's antibody, RNA-FISH (= using fluorescent-nucleotides binding to some RNA ), bacterial-CRISPR, siRNA, RT- PCR, northern-blot.
↑ This biology research did Not look into the detailed atomic mechanism, nor use the useless quantum mechanics, DFT, Alphafold-AI at all.
The 2nd and last paragraphs of this hyped news (7/3/2024) say
"An RNA switch is a synthetic mRNA molecule designed to regulate the translation of a gene in response to specific intracellular signals, such as microRNAs (miRNAs) or proteins."
"The split RNA switch offers a modular, scalable, and RNA-only platform for precise gene control, with strong potential (= just speculation, still useless ) for gene therapy, regenerative medicine, and synthetic biology."
↑ This research paper ↓
p.15~p.16-Methods used only macroscopic biological tools (= cells, RNA, antibody, natural enzymes.. ) without considering detailed atomic interaction nor using the (impractical) quantum mechanics, DFT.
↑ It is impossible to use biological tools without understanding the detailed atomic mechanism (= due to the useless quantum mechanical atomic theory ) for any practical purpose.
The 6th, 19th, 5th-last paragraphs of this hyped news (7/4/2025) says
"While browsing online protein databases, Kovalev spotted an unusual feature common to microbial rhodopsins (= based on natural organism's protein Not made by humans nor quantum mechanics ) found exclusively in very cold environments,"
"Our cryorhodopsins aren't ready to be used as tools yet (= still useless )"
"even a slight change in the position of a single atom can result in different properties"
↑ This research paper ↓
p.15-Fig.9B shows AI Alpphafold-3 prediction of static protein contains unreliable parts (= red ~ error ~ 50%, as in pIDDT score ) that can Not predict completely accurate proteins at a single atomic level nor exact biological (= dynamical ) function.
p.16~p.21-Materials and methods -- Not seeing atoms
used macroscopic biological tools and investigated static protein structures (= X-ray, crystal ) without considering atomic biological mechanism nor using the useless quantum mechanics.
The 1st-paragraph of this hyped news (7/3/2025) says
"A collaborative research team.. has identified microtubule (MT) destabilization as a promising (= still unrealized ) strategy to enhance platelet production from human iPS cell-derived immortalized megakaryocyte progenitor cell lines (imMKCLs)."
↑ This research paper p.3~p.5-materials and methods -- Not seeing atoms
used only macroscopic biological tools such as cell (= imMKCL ) culture, reagents, antibodies, flow-cytometry, in-vivo-mice, blood-platelet samples
↑ This stem cell research did Not look into atomic mechanisms (= so utilizing stem cell for curing diseases is impossible ) nor use the (useless) quantum mechanics, DFT.
The 10th, 2nd-last paragraph of this hyped news (7/7/2025) says
"Screening a set of drug compounds for growth-slowing effects in 32 different cell models of human cancers, they found that an experimental drug called rigosertib, which targets the MAPK pathway and is being tested against other cancer types, showed enhanced efficacy against ovarian cancer." ← Experiment or screening was needed instead of the (useless) quantum mechanical or AI predicting drug effectiveness.
"Dr. Hopkins hopes that these results will (= still unclear ) spur drug developers' interest in this approach (= this rigosertib failed in clinical trials in other diseases )"
↑ This or this research paper ↓
p.11-Limitations of this study says -- Still unclear
"the scale of these studies are
limited and thus do not capture the full breadth and depth of
ovarian cancer sub-types and disease states" ← still effectiveness is unclear.
p.17-p.19-Experimental methods -- No atoms, No quantum mechanics
used only macroscopic biological tools
such as cell culture, mice, drug screening, immunoblotting, immunochemical staining based on antibodies obtained from immunized natural animals without clarifying detailed atomic mechanism (= so cancers are incurable ) nor using the useless quantum mechanics, DFT.
See also this page.

Feel free to link to this site.