Top page (correct Bohr model including the two-electron atoms)
Back to protein structure page.
Strange "spin" is NOT a real thing
Black hole is an illusion.
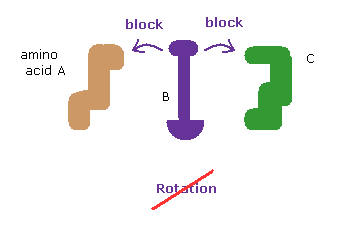
[Rotations are blocked in both directions. ]

The whole protein structure can be determined by dihedral (= torsion ) angles.
At each rotational sites, amino acids can be freely rotated to some degree (= rotation isomers. )
In these webpages, we show the collisions of two distant amono acids block these rotations and determine the whole protein structure.
So it is as if proteins act like rotating solid body.
Here we use the following sample JAVA program to investigate and manipulate protein structures.
Sample JAVA program ( PDB protein - rotation ).
This program can deal with up to about 500 amino acids per each subunit × 4. ( total 2000 amino acids. ).
"HETATM" parts (= other than usual proteins ) are incorporated into the former protein subunit.
First, we deal with following myoglobin.
Oxymyoglobin text ( 1MBO.txt ).
And this program can change each amino acid side chain except for proline.
So we also use the following text file as templates for 19 kinds of amino acids
19 amino acid templates ( tout.txt ).
(Fig.1) Change location folder.

Save this text at the location of "C:\sdff" folder as the filename of "1MBO.txt".
( Or you can save this protein text directly from PDB webpage. )
In the same way, save "tout.txt" in some folder, too.
( When you save in a different folder, change Fig.1 part inside program. )
After it, save the above sample JAVA program as "amic.java", and compile it.
You can run the program, as it is, neglecting notes such as "Recompile with -Xlint: unchecked for details ".
(Fig.2) Protein screen.
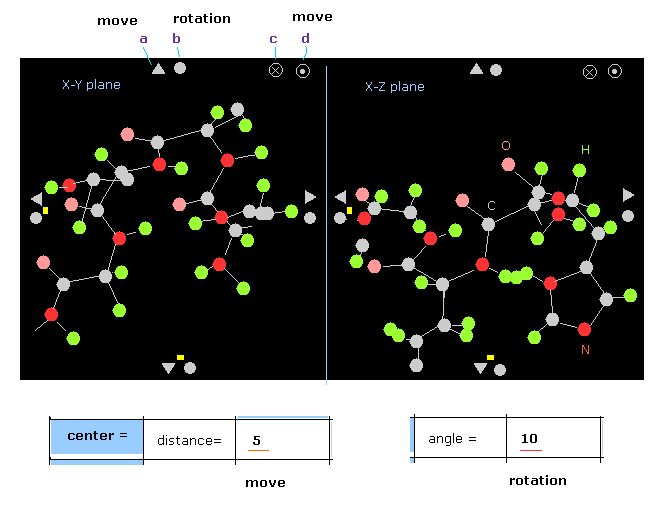
Basic manual is almost same as this page.
When you click some triangle (= a ), all protein moves in that direction.
The moving length per one click is based on textbox next to "distance =".
In Fig.2 case, if you click upper triangle, the protein moves 5.0 Å in +y direction.
( When this value is greater than 10, moving distance becomes 2.0 Å )
When you click some circle (= b ), the whole protein rotates in that direction.
The rotation angle per one click depends on the texbox next to "angle =".
In Fig.2 case, the rotation angle is 10 degree.
If you click "x " mark (= c ) on x-y plane, all molecules move in -z direction.
( When you click "point" mark (=d ) on x-y plane, they move in z direction. )
(Fig.3) Command prompt screen.
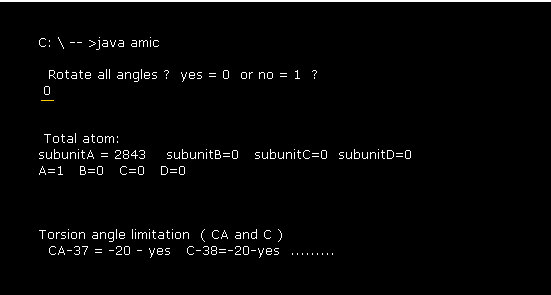
After running above program, the message of Fig.3 is displayed on command prompt screen.
When you input "0", this program investigates all dihedral angles ( CA and C), which is explained later.
Total atom's number of this 1MBO protein is "2843" including additional hydrogens .
1MBO consists of one subunit (= only A ).
(Fig.4) Text screen.
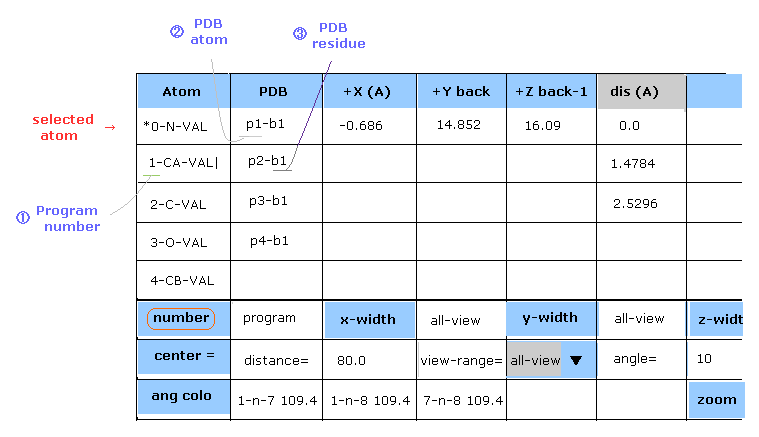
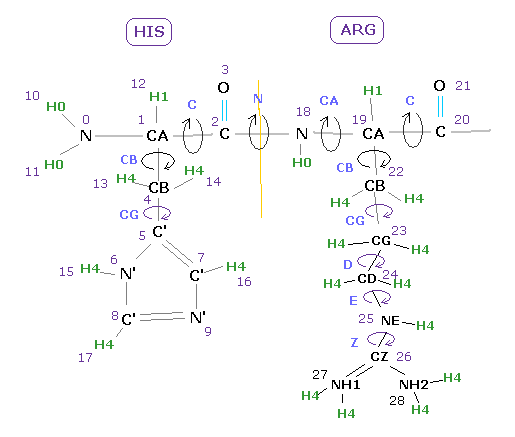
In this program, three atomic numbers are used.
The first is ( program ) atomic number, which is given to all atoms including hydrogen.
The second is PDB atomic number, and the third is PDB residue (= aminoacid ) number.
( Hydrogen atoms don't have PDB atomic number. )
(Fig.5) PDB data file ( example ).

If you click "number" button, you can change the kind of number displayed on the screen.
( Personally, PDB residue number is easy to see. Try it. )
Character "B" is added to B subunit atoms.
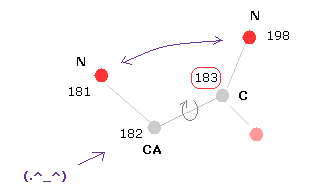
(Fig.6) Dihedral ( torsion ) angle.
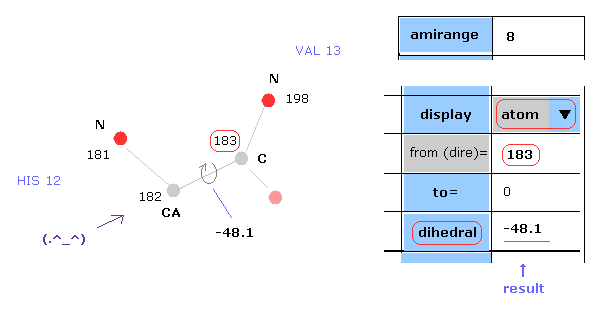
Dihedral ( torsion ) angle is a very important concept, because you can construct protein 3D structure only from the informations about all dihedral angles.
Dihedral angle means the angle between plane A (= 181-182-183 ) and plane B ( 182-183-198 ) in Fig.6.
When you input "183" into the textbox next to "from (dire)=" ( after choosing "atom" in scroll-bar ) , and click "dihedral" button, you can get dihedral angle of this ( between 182 and 183 ).
Here designate program number when "atom" condition, and PDB atomic number when "PDBat" condition.
(Fig.7) Choosing residue + "C".
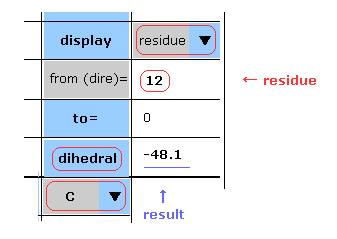
You can get dihedral hangles after choosing ( N, CA, C, CB, CG, ... ) and "residue" in each scroll bar.
In this case, choose "C", input PDB residue number (= HIS-12 ) into textbox, and click "dihedral" button.
( Basically, peptide bond is double-bond like, so in "N" case, this angle is about -180 degree. )
You need to be careful that hydrogens and oxygens are NOT involed in this dihedral angle at all.
(Fig.8) Dihedral angle and choice of atoms.
For example, if you want to know the angle between 18-19 (= before CA ) input atomic number of "19" into textbox, and click "dihedral" button.
Basically the angles just before CA and C carbons are referred as Ramachandran plot, which is very important for alpha helix and beta sheet structure.
If you input "4" (=CB ), you will know the angle between plane 0-1-4 and plane 1-4-5 in Fig.8 case.
If you input "26" (=CZ ), you will know the angle between plane 24-25-26 and plane 25-26-27.
( Of course, when you choose "residue" and "Z" in each scroll bar, and input residue number, you can get "CZ" dihedral angle. )
(Fig.9) Dihedral angle and atomic positions

As shown in Fig.9, when the atoms of 29 and 46 are arranged at "cis" positions, dihedral angles becomes "0" degree.
On the other hand, when they are at "trans" positions, the dihedral angle becomes -180 (or 180 ) degree.
From the viewpoint of Fig.9 (= arrow 30 → 31 ), vector 31-46 turns counterclockwise some angle ( for example 30 degree ), the dihedral angle becomes negative ( -30 degree ).
On the other hand, when it turns clockwise, its angle becomes positive.
(Fig.10) Distances between atoms on either side of C-183.
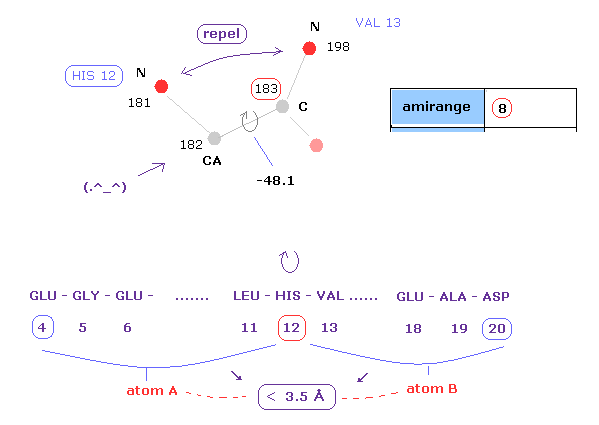
The important point is what determine each dihedral angle.
We pay attention to repulsive relations between atoms ( A and B ) on either side of designated dihedral angle.
When you click dihedral angle, after inputting "8" into the textbox next to "amirange", this program computes all distances ( excluding hydrogen ) between residue "4" (= 12 - 8 ) and residue "20" (= 12+8).
Here, atom A is inside residues from 4 to 12, another atom B is inside residue from 12 to 20.
Carbon atoms 182 (=CA ) and 183 (= C ) don't influence this dihedral angle at all.
( For example, the distance between 182 and 198 doesn't change by rotating this dihedral angle. )
In these cases, atoms 182 and 183 are excluded from this calculation.
(Fig.11) Distances less than 3.5 Å between two atoms on either side of 182-183.

This program display only interatomic distances less than 3.5 Å excluding hydrogen on command prompt screen.
As shown in this page, hydrogen atom has very week repulsive effects, so we neglect its effects here.
In Fig.11, for example, the distance between oxygen atom (= program number 130 ) and nitrogen (= 198 ) is 3.364 Å.
The shortest distance, which influences this dihedral angle is 2.765 Å, which is between nitrogens 181 and 198 of Fig.10.
The important point is that almost all distances are more than 2.8-2.9 Å due to steric hindrance.
( This is very important factor in determining protein structure. )
(Fig.12) Rotation of all amino acids after C-183 by 10 degree.
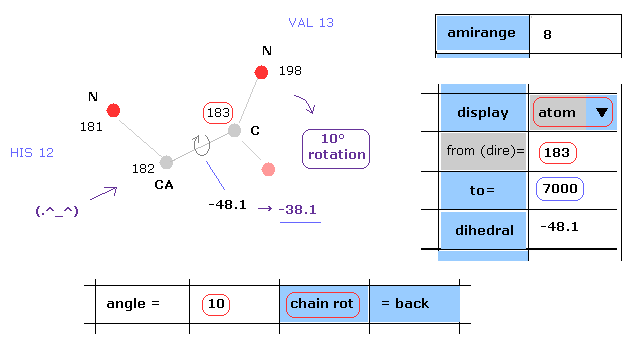
Next we rotate all amino acid chains after carbon-183 by 10 degree.
Input "183" and "7000" into textboxes next to "from(dire)=" and "to=".
( The last atom of myoglobin is 2843, so this "7000" can be changed into arbitrary number above 2843. )
And input "10" into the textbox next to "angle=", and click "chain rot" button.
This myoglobin includes 2843 atoms, so in this case, all atoms after carbon-183 are rotated around the axis of 182-183 atoms by 10 degree.
( So dihedral angle changes from -48.1 to -38.1 degree. )
(Fig.13) Rotation of all amino acids after C-183 by 10 degree - residue.
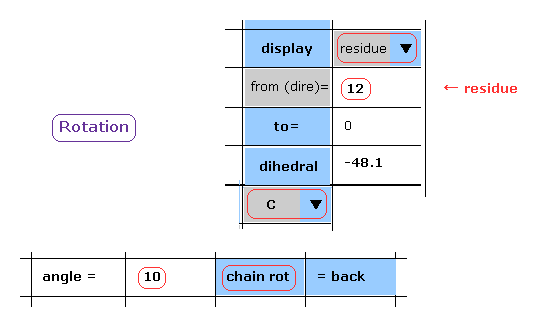
When you chooce "residue" and "C" in each scroll bar, you can rotate like Fig.12.
After input "10" (= rotation angle ) and "12" (= residue number at rotation site ) into each textbox, and click "chain rot" button.
In this case, irrespective of the value next to "to" button, all amino acids after C-183 are rotated by 10 degree.
(Fig.14) Interatomic distance change after rotation.

On the command prompt screen, interatomic distances, which become shorter than before rotation, are displayed.
As shown in Fig.14, the distance between nitrogens 181 and 198 becomes too short ( 2.765 Å → 2.690 Å ).
This site exerts strong repulsive effect, and blocks this rotaion by steric hindrance.
Shortest length before rotation is 2.764 Å, as shown in Fig.11.
So this shortest length becomes much shorter by this rotation.
(Fig.15) Rotation back → rotation by -10 degree.
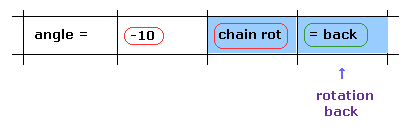
Next we investigate rotation in the opposite direction.
Click "=back" button to the original configuration.
Then input "-10" to the textbox and click "chain rot" button.
In this case, all amino acids after carbon-183 are rotated by -10 degree.
( As shown in Fig.9, positive angle is clockwise, negative angle is counterclockwise. )
(Fig.16) Rotation by -10 degree.

As shown in Fig.16, also -10-degree rotation, some interatomic distance becomes too short.
The atomic distance between oxygen 149 and nitrogen 214 becomes 2.698 Å.
In this case, alpha helix of these regions becomes too tight by this rotation.
So the dihedral angle of of -48.1 degree is the most stable state (= avoiding all obstacles ).
This means one or two conspicuous obstacles are key players to determine each dihedral angle.
(Fig.17) Most stable dihedral angle.

The key point is the shortest interatomic length.
Because even if there are only a few unreasonably short interatomic length, this rotation can be blocked.
So the present protein structure is the result of avoiding any single obstacle.
This is a true reason to determine protein struture.
(Fig.18) Almost all dihedral angles are fixed.
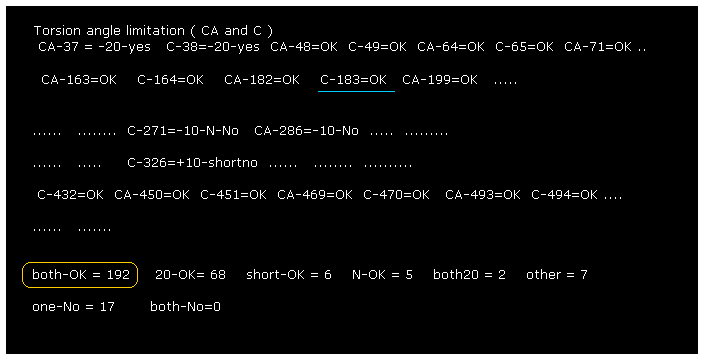
After running the program, if you input "0" (= Rotate all angles, yes ), the result of Fig.18 is displayed.
"CA-37" means rotations at program atomic number "37" (= CA ).
"both-OK" means the both-directional (= ±10 degree ) rotations are blocked like Fig.17, which means most stable.
"20-OK (yes)" means rotation in one-direction is blocked by +10 degree, and another direction is blocked by -20 degree.
So also in this case, both-directional rotations are almost blocked and stable.
"one-No" means one-directional rotation is blocked, but another-directional rotation is not blocked.
These sites are near surface of the protein, which rotation is probably blocked by other proteins or something.
The total number of both-blocked C, CA sites in myoglobin is 192, which is much bigger than 17 (= one is not blocked ).
This result shows even one or two obstacles (= shortest interatomic length ) are very important key factor to determine each dihedral angle.
(Fig.18') Rotations are blocked in both directions.

The whole protein structure can be determined by dihedral (= torsion ) angles.
At each rotational sites, amino acids can be freely rotated to some degree (= rotation isomers. )
As shown in this page, we show the collisions of two distant amono acids block these rotations and determine the whole protein structure.
So proteins act like rotating solid body.
(Fig.19) Rotation at C sites.
In C sites, the repulsive force between two nitrogens near C is very important.
+10 rotation makes this distance much shorter, so it is blocked.
-10 rotation makes this distance longer, but instead other atoms come too close to each other and blocked.
The calculation of this program considers only the shortest interatomic length.
In some cases, the original N-N length remains shortest after -10 rotation.
In these cases, if rotation is blocked by excluding only this N-N length, this case is displayed as -10-N-No.
And in some sites, we rarely see unnaturally short atomic length ( about 2.650 Å ).
Even when some rotations are not blocked, if they are blocked by excluding only this unnatural short length, words such as "+10-shortno" are displayed.
Try rotating these sites and confirm them ( program atomic number is displayed. )
(Fig.20) Changing amino acids regions.

In the above section, we compute only ±8 amino acids regions, as shown in Fig.10.
But there are some amino acid parts which distances are very close to each other, though their residue numbers are apart from each other.
Change the value of "int searchrange=20" in the above program, and run it.
This variable means computing amino acid range.
When this value is 20, this program compute all interatomic distances within ±20 amino acids on either side.
( Or use the program below. )
Sample JAVA program ( computing range ±20 ).
(Fig.21) Rotations are almost all blocked (= both-OK ).
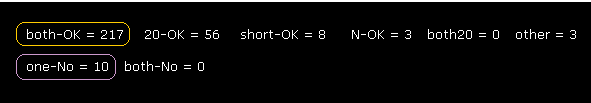
As shown in Fig.21, when we expand computing regions, we easily find that rotation of each dihedral angle is clearly blocked.
"both-OK" means rotations in both directions are blocked.
( In both ±10-degree rotations, the shortest interatomic length becomes much shorter = both rotation blocked. )
Total number of both rotation blocking sites is 217, which is much bigger than non-blocking sites (= only 10 ).
(Fig.22) Two distant amino acids crash.
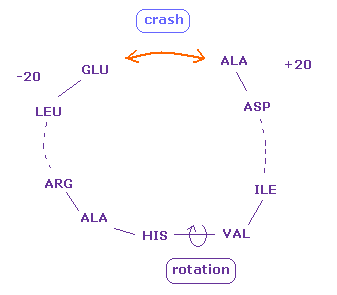
Even when residue numbers are apart from each other, original interatomic distances may be short.
In these cases, by rotating some sites which are apart from them, two atoms may crash into each other, like Fig.22.
As a result, "192" of Fig.18 (= ±8 range ) is increased to "217" in Fig.21.
These results indicate protein's structure is like some solid body.
When only a part of these solid bodies crash into each other, this conformation change is inhibited.
Even a few very short interatomic distances play important roles in determining protein structure.
(Fig.23) Chaging alanine into phenylalanine.
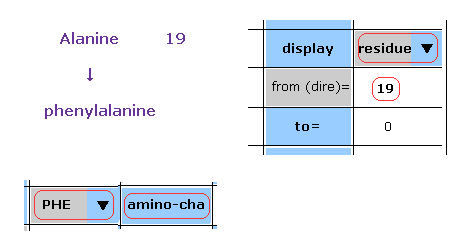
In this program, we can change the kind of each amino acid.
For example, residue 19 is alanine.
Choose "residue" in the scroll bar, input "19" into the textbox next to "from (dire)=".
And choose "PHE" in the scroll-bar ( seventh line ), and click "amino-cha" button.
(Fig.24) Alanine → Phenylalanine.
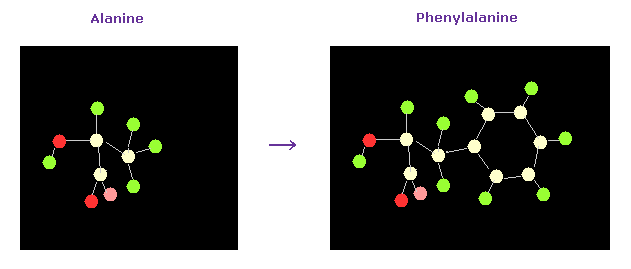
As a result, the designated amino acid is changed.
(Fig.25) Changing dihedral angle ( CB ) from 12 to 20 residues.
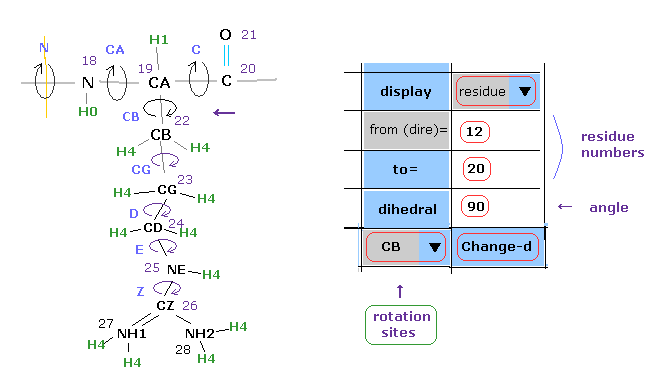
In this program, we can change dihedral angles of arbitrary positions included in some amino acid chains.
In Fig.25, we change all dihedral angle of CB site included in 12-20 amino acid residues.
( If amino acids without CB ( ex. GLY ) are included, these amino acids are automatically skipped. )
Choose "residue" and "CB" in each scroll bar, input the residue numbers of "12", "20" into textboxes, input
the resultant angle of "90" into the textbox next to "dihedral".
Then click "Change-d" button.
If so, all CB site dihedral angles in 12-20 amino acids are changed into 90 degree.
This manipulation is different from "chain rot".
In "chain rot", this rotation can be returned by "=back" button, but this "Change-d" cannot be returned.
(Fig.26) Amino acid construction.

This program can construct new amino acid chain.
Change the designated sites inside the program like Fig.26.
Sample JAVA program ( ami3.java protein construction ).
In Fig.26, change the variable like "int constr = 1".
"String sequence[]" mean amino acid chains we construct.
In Fig.26, 22 × leucines are constructed..
Change the variable "resn" to total number (= "22" ) of amino acids.
"CAang" and "Cang" mean the initial dihedral angles of all CA and C sites.
When CA and C dihedral angles are -60 and -50 degree, respectively, this protein becomes typical alpha helix.
(Fig.27) Hydrogen bonds in alpha helix (= total 22 amino acids ).
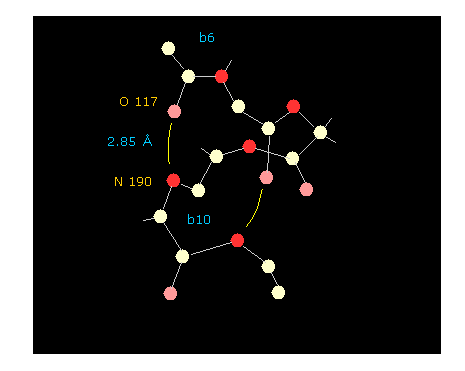
If you click "hydrogen" and "chain" buttons, you can display only main chain without hydrogens.
When you click some oxygen and nitrogen consequtively, interatomic distance between them is shown in the above textboxes.
As shown in Fig.27, oxygen of residue "6" and nitrogen of residue "10" form hydrogen bond, which length is as short as 2.85 Å.
If you change the whole dihedral angle by manipulation of Fig.25, you easily find how this hydrogen bond changes depending on dihedral angle. Try it.
(Fig.28) Range of fixed interaction in nitorgen.

As I explain in this page, when we compute this distance, we exclude fixed area like Fig,28.
For example, in case of nitrogen of Fig.28, interatomic distances between N and atom inside polygon doesn't change by chain rotation at C or CA.
This means other atoms outside this area determine each dihedral ( torsion ) angle near nitrogen.
(Fig.29) Interaction between N-3 and N-4.

In Fig.29, distance between nitrogen 3 and 4 ( N-3 and N-4 ) influences dihedral angle between them.
On the other hand, distance between N-3 and CA ( or C ) carbon doesn't change by this rotation.
So, in this case, CA and C atoms are inside fixed area and N-4 is outside it, as shown in Fig.28.
(Fig.30) Designating x-width.
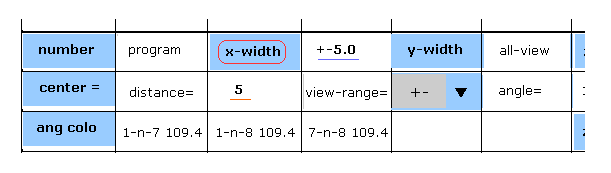
In this program, we can see cross sections of the whole protein.
One example is x-z (= left ), y-z (= right ) planes.
( Of course, in arbitrary planes, this operation is effective. )
Choose "+-" in the scrollbar of Fig.30, input "5" (= 5.0 Å ) into the textbox next to "distance=" and click "x-width" button.
In this state, only the designated area with the width of 5.0 Å in x direction is displayed in y-z plane (= right screen of Fig.31 ).
(Fig.31) Designating x-width.

If you choose "+" in the scrollbar, the area from 0 to +5.0 Å ( wrt. the mark ) in the x direction is shown.
( In case of "-", the area from -5.0 Å to 0 is shown. )
Caution: for example, x-z plane always displays all atoms in the direction of x and z.
So even if you limit "z" width in Fig,31, the screen doesn't change.
( Because, both x-z and y-z include z direction. )
When you choose "all-view" in the above scrollbar, and click x-width ( or y, z- width ) button, you can return to the original state.
If you click "number" button, you can change the kinds of atomic number displayed on the particle's screen below.
Basically residue number is short and eacy to see.
( And in PDB atomic number, hydrogen's number is not displayed, so this is also easy to see. )
(Fig.32) Change displayed atoms in textboxes.

If you click "Atom" button, next atomic column ( 5 - 9 ) are displayed.
If you click "+Z back-1" button, return to the column just one before that.
If you click "+Y back" button, return to the first atomic column (= 0 - 4 ).
If you click "+X (A)" button, each relative coordinate wrt. the selected atom is displayed.
"dis (A)" means the distance between each atom and selected atom.
( In this case, the distance between 0-N and 1-CA is 1.473 Å )
(Fig.33) Selecting atom.
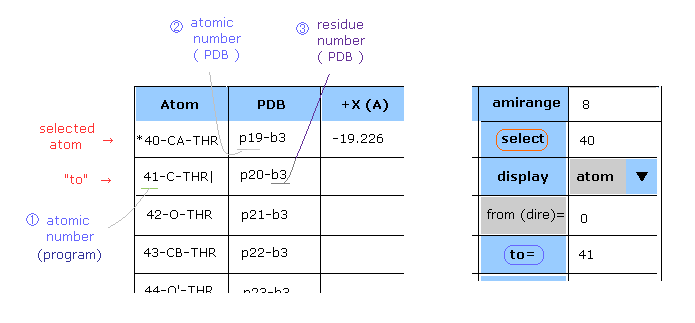
After choosing condition ("atom" or "PDBat") in the scroll bar, input atomic number into the textbox next to "select", and click that button.
In the case of Fig.33, (program) atomic number "40" (= carbon CA of threonine ) is chosen as "selected" atom. ( "*" mark is added. )
In "dis (A)" column, the distance between this selected atom and each atom is shown.
When you designate some atom inside B subunit using program atomic number, you need to add "10000" to atomic number.
( For example, atomic number "18" inside B subunit is "10018". )
In PDB atomic number case, you don't need to add this 10000, because PDB atomic number is specific.
In the same way, after choosing condition, input atomic number into the textbox next to "to", and
click "to" button, you can jump to the designated atom's textboxes.
" | " mark is added to this "to" atom.
This "to" atom is different from "selected" atom. This button is just to jump to that textbox.
In thse cases of Fig.33, when you choose "PDBat" in the scroll-bar, input "19" or "20" into each textbox instead of "40" or "41".
( Inside B subunit, you need to add "10000" to program atomic number. )
(Fig.34) Change "selected" atom by clicking atom directly on the screen.
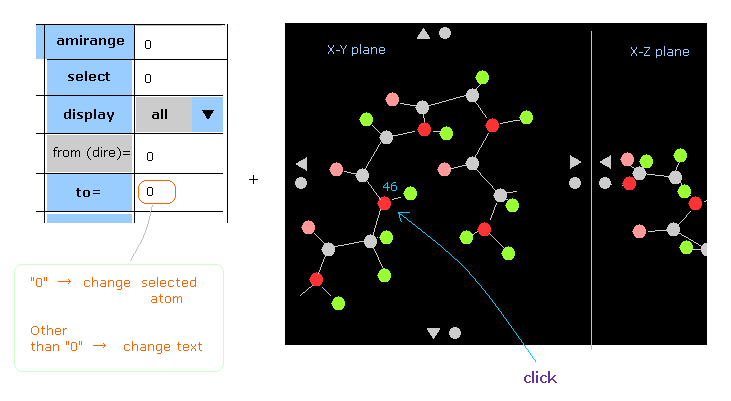
You can change selected atoms by clickling some atoms on the screen directly.
For example, click "46" red circle (= nitrogen ).
At this time, if "0" is inputted in the textbox next to " to= ", selected atom is changed into this "46" atom. "*" is added to it.
( You don't need to click " to= " button, just input "0" into the textbox before that. )
If you click "46" atom, in the condition that some values other than "0" is in this textbox, you only jump to text of "46" atom without changing selected atom.
( So in this case, mark " | " is attached to "46" atom. )
(Fig.35) "ang colo", "zoom", "x-y,x-z" buttons.
When you input some value into the textbox next to "zoom" and click that button.
The zoom of protein changes.
The value of "100" is initial state.
If you input "200", the whole protein is enlarged twice.
If you input "50", the whole protein is reduced to half of the original picture.
( First, you should reduce the whole protein to grasp the full-length picture. )
When you click "x-y,x-z" button, the left and right screen coordinates change.
The first is "x-y,x-z" planes, the second is "x-y, y-z" planes, and the third is "x-z,y-z" planes.
After clicking "ang colo" button ( see Fig.30 ), subunit A is red, B is green, C is pink, and D is yellow.
( And HETATM atoms become purple. )
So we can easily distinguish A and B protein subunits and the structural relation among them .
(Fig.36) "center" button.

After inputting some value ( for example "5" ) into the textbox next to "distance", and click "center" button, only atoms included in the area less than 5.0 Å apart from the selected atom are displayed.
The initial value is 80 Å.
If you combine this "center" and "width" buttons, various parts of proteins can be understood easily.
(Fig.37) Display distance between two arbitrary atoms.
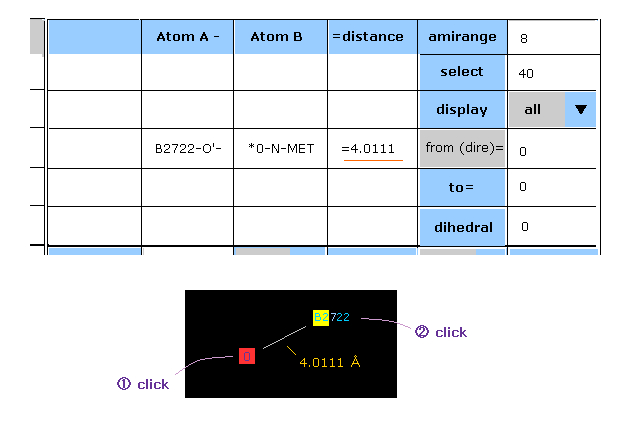
When you click two arbitrary particles consecutively, interatomic distance between them is shown in the textbox.
In Fig.37, first click N atom of methionine (= atomic number 0 ), and second click oxygen atom of H2O (= B subunit 2722 ).
These numbers are program atomic number.
In Fig.37 case, O atom is selected atom, which is displayed as a square.
And the atom clicked now is shown as "yellow square".
( Of course, when you also see "dis A" column, you can find the distance between each atom and selected atom. )
Other commands are almost same as this page.
You can choose only "residue" condition in the scroll-bar, when you use "from to" display command.
For example, if you input "1" and "5" into the textboxes next to "from" and "to", and then click "display" button, only aminoacids of subunit A with the residue number 1-5 are displayed on the screen.
When you input "10001" and "10005" into the textboxes ( "from" and "to" ), and click "display" button, only aminoacids of subunit B with the residue number 1-5 are displayed on the screen.
In case of "C" and "D" subunits, add "20000" or "30000" into residue number.
About the "dihedral", "chain rot" button, they are the same as this page.
When you know the dihedral angle inside subunit B, add "10000" to (program) atomic number.
( You don't need to add "10000" to PDB atomic number. )
And in this program, we can designate some site by residue + scroll bar, as shown in Fig.13.
Basically, one subunit protein ends at "TER".
The proteins following "TER" is the next subunit.
"HETATM" parts are counted as the same subunit, which is just before that.
(Fig.38) Turn the whole protein in some direction.
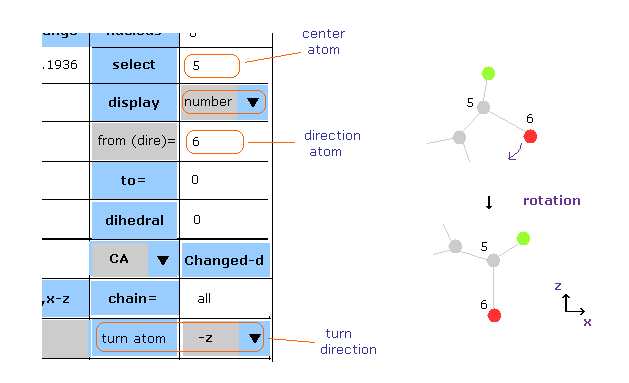
First we choose some atom as selected atom (= rotation center ).
Next, input some atomic number into the textbox next to " from (dire)= " ( after choosing condition "atom" or "PDBat" in the scroll-bar ), choose direction ( x, y, z, -x, -y, -z ) in the scroll-bar ( bottom line ), and then click "turn atom" button.
In Fig.38 case, the whole protein rotates to turn vector 5 → 6 atoms toward -z direction.
Of course, if you choose some atom inside B subunit ( not using PDB atom ), you need to add "10000" to program atomic number.
( So program atomic number "1" in B subunit → "10001". )

2013/8/29 updated. Feel free to link to this site.