

(Fig.1) Immunofluorescence, Western blotting in the current biotechnology just show some "stain" as a vague sign of attaching antibodies to proteins, which is useless without clarifying nor manipulating actual atomic interactions between proteins.
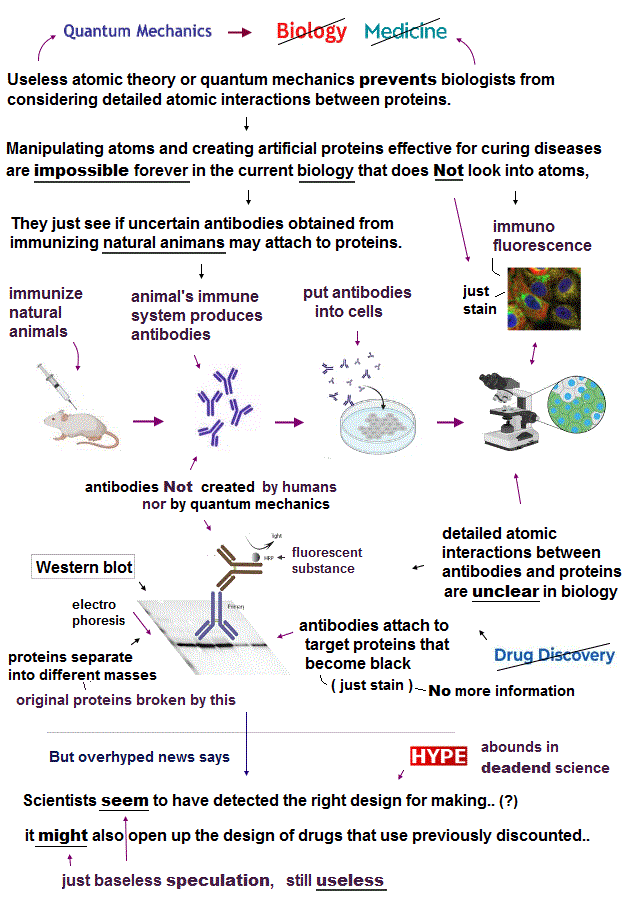
All tools used in the current biological and medical research such as PCR, polymerase, DNA-ligase, restriction enzymes, nucleotide-protein-immunofluorescence antibodies were obtained from natural bacteria, viruses or immunized animals, Not designed by human's technology nor calculating actual molecular interactions using (fictional) quantum mechanics (= Schrodinger equation ) which impractical quantum mechanical calculations are Not used in any molecular biology research.
↑ This fact shows the mainstream basic physics or quantum mechanics made No contribution to Biology, medical research or drug development ( this abstract-first, this 4,Biologists' view, this p.30-conclusion, this p.9-second-paragraphs~, this p.6-fifth-paragraph, this two arguments against quantum brain )
All the current molecular or protein simulating methods consisting of quantum mechanics, density functional theory (= DFT ) and molecular dynamics (= MD ) are unable to deal with actual proteins and enzymes.
So biologists and medical researchers are unable to utilize actual atomic interactions for considering biological reactions, which fact makes it impossible to design and create truly useful drugs without side effects (= predicting presice side effects definitely needs the knowledge of detailed atomic interactions ), which inconvenient truth of deadend science is often hushed up by an incredible amount of the media-hypes.
For example, the 6th and 9th paragraphs of this typical hyped news (or this ) argue
" Scientists seem (= still speculation ) to have determined the right design for making the drugs sufficiently safe and highly effective ?"
"And exploiting it could (= just speculation ) really open up the field for these drugs. Pharma companies now can imagine (= just imagine ) using them in a broader range of tumors than previously anticipated. Moreover, Lambert said, it might (= baseless speculation, again ) also open up the design of drugs ?"
Impossibility of utilizing atomic interaction based on practical atomic theory made biologists rely on natural organisms' enzymes and antibodies obtained from immunizing natural animals as the only tool to vaguely investigate proteins' intearctions (= instead of designing and creating artificial proteins or new original enzymes based on detailed atomic interaction ).
Immunized (natural) animals by some target proteins or antigens → collecting antibodies produced by natural animals' immune system (or getting animals' spleens or humans' B cells that may ambiguously include DNA encoding antibodies against injected proteins in monoclonal antibody production ) → Using those antibodies to see if they attach to some proteins without knowing detailed atomic interaction.
↑ These typical biological tools to just see if the natural-animal-based antibodies ( this p.10 ) may vaguely attach to target proteins are called immunofluorescence ( this = this p.13 ) and Western blotting (= WB = just seeing blurred stains with No more detailed information, this p.3, Fig.2, this = this Fig.1A ).
Other biological methods or names similarly using natural-animal-based antibodies attached to target proteins are immunoassay, immunoblotting ( this = this p.12 ), ELISA ( this p.3 ), flow cytometry ( this = this p.19, this p.14 ), immunoprecipitation ( this p.10, this = this p.21 ), immunohistochemistry ( this = this p.6 )..
↑ All of these biological methods just see "(immunofluorescence) ambiguous stain" or (Western) blotting which allegedly indicate (natural) antibodies attached to some target proteins, which antibodies do Not clarify detailed atomic interaction between antibodies and proteins nor manipulate target proteins for curing diseases.
Because in these immunofluorescence and Western blotting, biologists are unable to know how exactly or what atoms each antibody sticks to. ← How antibodies and target proteins change their conformations after binding (= this knowledge is neccesary for developing useful drugs ) is unknown in the current biology.
Even whether antibodies really stick to target proteins is unclear, as shown in unpredictable vaccine's effectiveness, side effects (= vaccine side effect is caused by antibodies binding to unexpected, wrong proteins ) and frequently-failed immunotherapy.
See other examples of biological researches using these "immuno-" or "Western"-blotting based on natural-animal-based antibodies without looking into detailed atomic interaction, this = this p.15-16.
Furthermore, the current immuno-blotting or Western blotting has to separate the original mixed proteins by SDS-PAGE electrophoresis which denatures and destroys the original proteins' structures and properties, hence, even if antibodies attach to such denatured proteins, it is likely to be unrelated to clarifying the actual natural proteins' affinities or properties.
↑ Without using these denaturing agents in the methods called native PAGE, many different native proteins remain sticking to each other, so distinguishing and identifying proteins by different masses become more difficult with lower resolution ( this p.2, this-intro-2nd-paragraph, this p.2-2nd-paragraph, this-6th-paragragh ), which means the current biotechnology cannot investigate or know the original proteins' activities correctly.
All the current methods and tools used in biotechnology and medicine are obtained from natural organisms irrelevant to (useless) quantum mechanical atomic theory, and unable to utilize detailed atomic interactions (based on practical atomic theory), which fact makes it impossible to develop effective drugs or treatment against deadly diseases.
PCR or polymerase chain reaction is one of most popular biological methods for increasing and amplifying target DNA genes after preparing short DNA primer genes (= attach to both edges of target-protein genes ) by using enzymes called polymerase originating from natural bacteria (or natural polymerase whose gene is slightly modified using other natural enzymes ).
↑ No quantum mechanical methods are involved in discovering or developing PCR enzymes.
RT-PCR or reverse transcription polymerase chain reaction is the method of detecting RNA genes such as COVID-19 viruses by changing the target RNA into complementary DNA (= cDNA ) and amplifying those cDNA using ordinary PCR.
The enzymes called reverse transcriptase used in this RT-PCR were derived from natural retroviruses (or their slightly-gene-modified enzymes), Not from being designed or created by (useless) quantum mechanical calculation.
Real-time PCR is the method of quantifying the target DNA genes by using ordinary PCR and fluorescent molecules. ← All of these biological techniques are developed based on actual experience and experimental observations, and No quantum mechanical prediction is involved.
Sequencing is a technique used for reading and determining the order of nucleotides (= adenine-A, guanine-G, cytosine-C, thymine-T ) in the target DNA or RNA.
DNA sequencing uses the ordinary DNA polymerase and "stop-nucleotides labeled with different-color fluorescent molecules (= A, G, C, T nucleotides are labled in different colors ), measure the random lengths of randomly-amplified DNA fragmants by electrophoresis, identify what nucleotide is at the edge of each DNA fragment by measuring colors, which eventually could determine the whole target DNA's sequence or order of nuleotides.
No quantum mechanical calculation is involved in this technique.
RNA-sequencing is also for determining RNA nucleotides by changing the target RNA into cDNA by reverse transcriptase and using the method similar to DNA sequencing.
The examples of researches using this sequencing are this = this p.11
DNA microarray is a technique to see if target DNA can bind to some complementary nucleic acid sequences based on experimentally-known complementarity ( A-T, G-C ), not on quantum mechanics.
Small interfering RNA (= siRNA ) is a short nucleotide RNA molecule used in silencing or supressing the expression of the target genes by binding to target nucleotides through the well-known RNA complementarity, Not by quantum mechanical calculation.
This siRNA or gene silencing utilizes the mechanisms inherent in natural organisms, whose silencing mechanism was Not created or designed by human technology from scratch or useless quantum mechanical calculations.
Short-hairpin RNA or shRNA is also one of these siRNAs that utilizes natural organisms' gene silencing mechanism and enzymes.
To roughly know the amount of DNAs and RNAs, the absorbance of lights with some specific wavelengths are used.
Rough determination and quantification of total protein's concentration also uses the similar measurement of light absorption called Bicinchoninic acid assay.
↑ All these methods are unable to determine precise structure or identify the target protein, and these techniques are based on empirical observations, Not on (useless) quantum mechanical calculations.
Mass spectrometry (= MS ) is the method of determining proteins' masses by ionizing target proteins and fly them under electric field where heavier proteins move slower (or harder to accelerate ) than lighter proteins. which is obvious in classical mechanics. Quantum mechanics is unnecessary.
This MS just estimating masses cannot know the detailed protein interactions (ex. this = this p.15-right, this = this p.7-9 ).
SDS-PAGE is an electrophoresis method that separates proteins by masses whre bigger proteins move slower than smaller proteins, which is common sense by classical mechanics.
This SDS-PAGE or electrophoresis cannot know the protein's detailed structure or interaction. ← inapplicable to developing drugs
Protein chromatography separates proteins by their different sizes, different affinities with different ligants or antibodies or light scattering based on experimental observations and general knowledge.
No quantum mechanical calculations are involved.
Proteosome assay roughly measures the activity of natural proteasome enzymes by using substrate and fluorescence ( this p.3 ), based on empirical observation, Not on (useless) quantum mechanical calculations.
EMSA (= electrophoretic mobility shift assay ) is used to see if DNA roughly binds to proteins by using electrophoresis where proteins bound to DNA become heavier and move slower, which is common sense.
This method cannot know the detailed interaction mechanism between DNA and proteins.
Electron microscope, Cryo-EM, X-ray crystallography, and atomic force microscopy can determine the protein's structure even in the atomic-level resolution.
But the current unrealistic quantum mechanical atoms, its useless density functional theory (= DFT ) and (pseudo-classical) molecular dynamics prevent scientists from using real atomic picture for understanding real protein interactions even after using these high-resolution microscopy.
↑ Useful drug development is impossible forever, as long as we rely on the unrealistic quantum mechancial atomic model.
ATP assay is a method of roughly estimating the amount of natural molecules called adenosine triphosphate (= ATP ) by using luciferase enzymes originating from natural organisms and measugin emitted light.
CRISPR-Cas9 system used in gene editting was obtained from natural bacterial immune systems, Not from being artificially designed or created by humans or useless quantum mechanical calculations.
This research also used the flow-cytometry based on natural-animal-based antibodies ( this p.3 ) without looking into detailed atomic interaction.
Synsthesis of mRNA (= messenger RNA ) by DNA → RNA transcription in vitro uses enzymes called RNA polymerase originating from natural organisms such as bacteriophages (e. T7 bacteriophage or T7 phage ) that are natural viruses infecting bacteria or Escherichia coli ( this introduction ) combined with natural nuleotides such as ATP, CTP, GTP.. ( this p.5, this = this p.9-right ).
These enzymes were Not designed or created by humans nor by (useless) quantum mechanical calculation.
The recent research on brainstem neutron uses the green fluorescent protein or GFP isolated from natural jellyfish whose gene was inserted into plasmid vectors found in natural bacteria (= that was injected into mouse brainstem ) and immunostaining based on antibodies obtained from immunizing natural animals (= this p.10-AAv-injection and optical fiber implantation, p.11-immunostaining ). ← No atomic interaction was considered, so No effective drug discovery. No (useless) quantum mechanical DFT nor molecular dynamics is involved.
Overhyped-AI cellular biology also uses only the natural-organism-based tools such as plasmid, GFP, immunofluorescence, Western-blot based on natural-animal-based antibodies without considering any atomic interaction (+ No useless quantum mechanics, DFT, or MD is used, this p.16-17 ), so useless for practical drug discovery.
The recent research on organoids also relied only on immunohistochemical staining using natural-animal-based antibodies, microarrays, RNA sequencing (= this p.23- Histological analysis, Tissue microarray. p.24- sequencing ), all of which are unrelated to (useless) quantum mechanical calculation (= slimilar research is this = this p.24 ).
This research (= this p.6-method ) also used only immunoprecipitation based on natural-animal-based antibodies and liquid-chromatography (= LC ), mass spectrometry (= MS ) based on ordinary classical mechanics (= heavier proteins move slower ), which did Not look into atomic interaction (= so finding effective treatment for Alzheimer is impossible ) nor use (useless) quantum mechanics.
This research (= this p.24-27 ) about breast cancer metastasis used the ordinary immunofluorescent staining, immunoblotting, immunoprecipitation based on natural-animal-based antibodies, cloning some DNA into (natural) plasmid vector by RT-PCR, quantitative PCR using natural-organism-derived enzymes, mass spectrometry, liquid chromatography based on common sense (= heavier protein moves slower ), which does Not use (useless) quantum mechanics nor clarify the detailed atomic-based protein-protein interaction, so it will never lead to developing effective cancer treatment (= instead, they always just used the misleading vague word "potential", this-last-paragraph ).
This research (= this p.11-materials and methods ~p.15 ) also will never lead to effective cancer therapy, because they did Not look into any detailed atomic interaction due to useless quantum mechanics that prevents creating really-useful artificial proteins, instead, they rely on natural-organism-based plasmid-vector's DNA construction, CRISPR-Cas9, RNA-sequencing, immuno-blotting, immuno-precipitation, flow cytometry based on natural-animal-based antibodies.
This brain cell's research (= this p.10-experimental-p.12 ) also just used natural organisms' proteins and mechanisms such as (jellyfish) GFP fluorescent proteins, (bacterial) plasmid vector, shRNA, Western blotting based on natural antibodies without looking into any detailed atomic interaction due to useless quantum mechanics.
This reserach (= this p.11-Method-p.12 ) on allegedly-molecular insight into leaflet also just used ordinary macroscopic biological tools derived from natural organisms or enzymes such as (bacterial) plasmid construction, RT-PCR, gene-sequencing, EMSA, without looking into detailed atomic interaction nor using (useless) quantum mechanics.
Similar recent researches using only natural-organism-based antibodies, enzymes, cloning, constructing of viral plasmid vector's recombinant DNA without using (useless) quantum mechanics nor looking into detailed atomic interactions are this = this p.2-14, this = this p.13-14, this = this p.3-9, this = this p.10-16, this = this p.2-method~p.4, this = this p.12-20, this = this p.33-40, this = this p.2-8, this = this p.11-method-p.13
↑ Researchers are doing these very time-consuming fruitless works only for aiming to publish their papers in top journals instead of really curing diseases.
The present biological and medical reseaches lacking presice atomic mechanism are meaningless, just waste of time ruining researchers' future as slaves to greedy academic journals.
The 1st, 4th and 10th paragraphs of this recent news hyping Alzhemer's research say
"While newly approved drugs for Alzheimer's show some promise for slowing the memory-robbing disease, the current treatments fall far short of being effective at regaining memory (= Alzheimer is incurable in the current impractical mainstream science just wasting time for aiming at publising papers in journals instead of really aiming at curing diseases )."
"Research has shown that KIBRA (= protein ) is required for synapses to form memories, and team has found that brains with Alzheimer's disease are deficient in KIBRA."
"Interestingly, KIBRA restored synaptic function and memory in mice, despite not fixing the problem of toxic tau protein accumulation (= root cause of Alzheimer is still unknown ). Our work supports the possibility that KIBRA could (= just uncertain possibility, still useless ) be used as a therapy to improve memory after the onset of memory loss, even though the toxic protein that caused the damage remains (= toxic proteins cannot be reduced )"
↑ This medical research just vaguely saw the existence of various proteins such as KIBRA, tau and synapse by the methods of immunoblotting, Western blotting, immunofluorescence using antibodies obtained from immunizing natural animals without looking into detailed atomic interaction (= so precise atomic mechanism of Alzheimer is unknown ) and without (useless) quantum mechanical calculations.
↑ This p.16-left-1st-paragraph-immunolabel by antibody, 2nd-paragraph-Western blot by antibody, p.16-right-1st-paragraph-immunoblot, 2nd-paragraph-immunostaining by antibody, p.17-left-1st-paragraph-antibody, 2nd-paragraph-Western blot (= by antibody ).
↑ Just vaguely seeing immunostaining or Western blotting by uncertain antibodies can Not tell us true cause of Alzheimer diseases (= at precise atomic or molecular level ), hence, finding effective drugs (= which needs the knowledge of precise atomic interaction between drugs and proteins ) is impossible in the current medical research hampered by impractical basic physics or quantum mechanics.
The 1st, 7th paragraphs of another recent hyped news say
"Researchers.. have unveiled unprecedentedly detailed images of brain cancer tissue through the use of a new microscopy technology called decrowding expansion pathology (dExPath).Their findings, provide novel insights into brain cancer development, with potential (= uncertain future, still useless ) implications for advancing the diagnosis and treatment of aggressive neurological diseases (← ? )."
"Antibodies, however, are large and many times cannot easily penetrate cell structures to reach their target. Now, by pulling proteins apart with dExPath, these same antibodies used for staining can penetrate spaces to bind proteins in tissue that could not be accessed before expansion, highlighting nanometer sized structures (= but Not looking at actual atomic interaction needed for clarifying true mechanism of diseases ) or even cell populations."
↑ They just used the methods of immunostaining that can vaguely see whether fluorescent antibodies bind to target proteins without looking into detailed atomic structure, hence, this biological research is unable to clarify true cause of brain diseases and unable to cure cancers.
This p.2-antibody, immunostaining, p.18-46-immunostaining, p.50-51-antibodies obtained by immunizing natural animals. ← Antibodies were Not designed or created by humans nor (useless) quantum mechanical technology.
The 1st and 7th paragraphs of this hyped news say
"A research.. has succeeded for the first time in identifying at the molecular level the dynamic mechanism used by the enzyme RNase R to degrade the ribosomal 30S subunit (← actually, molecular mechanism has Not been elucidated or considered in this research )"
"The researchers knew from previous work that certain enzymes, such as ribonuclease R (RNase R), are involved in the degradation process of ribosomes in stress situations. Using cryo-electron microscopy, they were able to show for the first time that the enzyme RNase R binds to the small 30S subunit of the ribosome." ← No mention of practical use
↑ This research observed the protein structure of ribosome and RNase using cryo electron microscopy (= Cryo-EM ), which is one of electron microscopes observing frozen samples at extremely low temperature, but they could Not explain molecular mechanism of this protein interaction due to useless basic quantum mechanics.
This p.9-methods used plasmid and immuno-precipitation just vaguely seeing whether antibodies bound to target proteins without looking into detailed molecular interaction.
This p.10 says nothern blots, western blots (= just vaguely seeing whether antibodies or nucleotides bind to target sample ), cryo-EM, and Alphafold that is based on experimentally-observed protein structure database (= PDB, this p.2-right = No quantum mechanics is used ) and do Not consider actual molecular interaction, so useless for drug discovery.
This p.11 used mass spectrometry (= of protein ), RNA sequencing, immunoprecipitation. ← No (useless) quantum mechanics nor atomic interaction was used in this biological research.
↑ This means biologists just observed protein structure by microscope and cannot utilize those observed results (= so waste of their time ), because they cannot access actual atomic or molecular interaction necessary for drug development (= due to useless quantum mechanics ), which is why the current biology or medicine is deadend. Deadly diseases such as cancers and Alzheimer are still incurable.
One of the reasons is when researchers try to explain proteins or ribosomes from practical microscopic atomic or molecular viewpoint, academic journals always require them to rely on unrealistic impractical one-pseudo-electron DFT model (= lacking real atomic or molecular shapes ) or unrealistically-time-consuming molecular dynamics (= MD, this p.5 ), which useless mainstream methods are why the biological or medical researchers can Not utilize actual atomic or molecular interactions to explain protein or enzymatic reaction, as shown in this latest research.
The 1st, 4th, 8th, 12th, last paragraphs of this hyped news say
"But, it's much harder to provide cells with instructions on how to organize and use those new parts. Now, new tools from University of Wisconsin–Madison researchers offer an innovative way around this problem (= untrue, this research can Not manipulate proteins or molecules inside cells at all )."
"for example, the proteins MinD and MinE—known collectively as MinDE—interact with each other along the cell membrane to produce wave-like patterns, which aid in the movement of molecules within the cell (= these MinD-and-E are natural proteins of bacteria involing in cell division, and showing some oscillation, their detailed functions are still unknown, this summary )."
"Biochemists at UW–Madison have developed a tool to control movement and organization of specific proteins in mammalian cells (= actually, they did Not control proteins at all in this research that just measured fluorescent light ) while leaving other proteins alone. Their new tool harnesses the waves and oscillations derived from interactions between MinDE proteins, which are found only in bacteria and do not interfere with mammalian cellular function."
" As each ratio of MinDE proteins emits a unique oscillatory pattern, the proteins can be inserted into a group of cells to give each cell its own pattern—an individual beacon that lets researchers observe each cell more easily."
"The Coyle Lab plans to continue (= which phrases mean "still useless research now" ) exploring the tool's applications, including studying the dynamics of signaling pathways in tumors.."
↑ This research ( this p.2-3 ) just used fluorescence microscopes and observed various colored lights emitted from natural fluorescence proteins such as jellyfish-GFP (= green fluorescent protein ) fused with bacterial proteins Min-D and Min-E by connecting those genes in lentivirus vectors (= using enzymes or viruses derived from natural organisms, Not from useless quantum mechanics ), which infect the cultured mammalian cells where bacterial proteins of Min-D and Min-E were produced, oscillating and emitting light due to fusing with fluorescence proteins.
↑ So this research did Not look into each protein or molecular interaction, much less controlled nor programmed protein activity ( due to useless quantum mechanics ).
All they did was just observe light emitted from fluorescent proteins ( this-middle-section snippets ).
This p.2-Abstract says
"MinDE circuits (= oscillating bacterial proteins ) act like “single-cell radios”, emitting frequency-barcoded fluorescence signals that can
be spectrally isolated and analyzed using digital signal processing tools (= fluorescence microscope )."
↑ Finding new effective drugs or treatments is impossible in the current biological or medical research looking away from atomic interaction.
This research paper ( this p.2-2nd-last-paragraph ) says
"We used lentivirus to express mCherry-MinD and MinE-GFP in multiple different mammalian cell lines (= bacterial proteins MinD or E are fused with fluorescence proteins mCherry or GFP in lentiviral vectors that can infect cells which eventually express these proteins ) and
imaged their spatiotemporal distribution by timelapse fluorescence microscopy ( this Fig.1~4, MinD protein is colored red by mCherry, and MinE protein is colored green by GFP )"
This p.5-4th-paragraph says
"Because MinDE (= bacterial proteins ) circuits are fast and protein-based, more complex signaling dynamics or composite signals
can be created by coupling MinDE to other proteins or cellular structures..
When FRB (= one of natural proteins ) was fused to MinD, an FKBP-BFP (= another protein + fluorescence protein ) test payload did not colocalize
with MinDE in the absence of rapamycin. However, within seconds of adding rapamycin, the BFP signal
colocalized with MinDE in space and time. As a result, this circuit generates a new oscillatory
signal in the BFP channel whose frequency matches the MinDE carrier signal."
↑ They fused natural protein called FKBP with another fluorescence protein BFP (= originating from sea anemones ), and fused MinD (= an oscillating bacterial protein bound to red mCherry fluorescence ) with another natural protein called FRB (= FKBP-rapamycin binding domain ).
↑ When they put rapamycin (= adhesive molecule derived from bacteria ) into it, the rapamycin connect both FKBP and FRB proteins, and resultantly, the oscillations of two fluorescence proteins from MinD-mCherry and FKBP-BFP are slightly changed ( this p.6-Fig.3 ).
↑ Thye call it "programming (= actually far from programming you imagine )", but they could Not control each protein nor know exact protein or molecular intreaction. All they could do was observe the oscillating fluorescence lights from microscope without looking into detailed protein's structure or interaction.
This p.6-last~p.7 says
" For this circuit, we fused a substrate sequence for PKA (= natural protein kinase A ) to MinD (= bacterial protein ) and co-expressed this with mCerulean-FHA (= natural protein + fluorescence ), which specifically binds to the phosphorylated substrate sequence.. The resulting
MinDE circuit generates a new signal derived from MinDE in the mCerulean channel whose amplitude is
dynamically modulated by PKA signaling activity"
↑ In this case, they fused natural protein called FHA (= Forkhead-associated domain ) with another fluorescence protein called mCerulean (= originating from Aequorea victoria ), and fused MinD (+ mCherry fluorescence ) with PKA kinase (= natural protein phosphorylating ) substrate where activated PKA (= protein kinase ) phosphorylated PKA-substrate, connected it with FHA protein, which changed the oscillating patterens of lights emitted from those two fluorescence proteins ( this p.7-Fig.4 ). ← detected by fluorescence microscope
↑ This research just observing fluorescent light did Not look into nor consider detailed atomic interaction between proteins, so controlling proteins (or finding effective treatments or drugs ) without knowledge of actual protein or molecular interactions is impossible.
The current biological research heavily relies on these fluorescence proteins such as jellyfish-GFP, but fusion with these fluorescence proteins could cause serious artifact, change or destroy the original protein's properties ( this 2~3rd paragarphs, this p.2-left-2nd-paragraph ), so it is impossible to know the true protein mechanism by the current biological method.
Of course, the useless quantum mechanical calculations such as one pseudo-electron DFT or extremely-time-consuming molecular dynamics were Not used in this biological research.
This biological research neglecting detailed atomic interaction shows researchers are obsessed only with publishing papers in journals instead of really aiming at curing diseases.
This recent study tried to find artificial enzymes that could break stubborn silicon-carbond bonds in widely-used chemicals known as siloxanes by time-consuming trial-and-error approach without looking into detailed atomic intearction, and failed.
The 4th, 13th, 15th, 19th paragraphs of the same news say
"The researchers say that while practical uses for their engineered enzyme could still be a decade away or more,.. (= still useless, they failed to create practical enzymes after all. )"
"In the new study, the researchers wanted to find ways to break the bonds rather than create them. The scientists used directed evolution to evolve a ( natural ) bacterial enzyme called cytochrome P450 (= based on natural-organism's enzymes instead of ones designed and created by humans or useless quantum mechanical atomic theory )."
"They mutated the DNA of the cytochrome P450 (= using natural organism-based enzymes as biological tools ) and tested the new variant enzymes. The best performers were then mutated again, and the testing was repeated until the enzyme was active enough to enable the researchers to identify the products of the reaction and study the mechanism by which the enzyme works (= time-consuming trial-and-error- approach without looking into detailed atomic interaction )."
"The final improved enzyme does Not directly cleave the silicon–carbon bond (= after all, this trial-and-error approach could Not create the desired artificial enzyme )".
↑ To create desired artificial enzymes such as killing cancer cells without side effect, we need to consider actual atomic interactions using practical atomic model (= which is obstructed by the current useless mainstream quantum mechanical pseudo-models ) instead of the extremely-time-consuming blind trial and error approach used in the current mainstream biological and medical research.
The recent "engineering bacteria" research also just slightly modified genes of natural proteins of ferritin ( this p.2 ) and polyhedrin, constructed plasmid DNA (= circular DNA found in natural bacteria or various organisms ) by using enzymes obtained from natural microbes, instead of truly designing and engineering new innovative proteins ( this 5-7th paragraphs = this p.3 ). ← No atomic interaction is considered, so Not clarifying detailed mechanism of proteins, and unable to develop new really-useful proteins
Artificial cells just encapsulate natural proteins such as myoglobin (= Mb ), β-Gal, actin, calcium phosphate, ALP ( this p.12 ), which is Not truly artificial cell nor creating new artificial proteins.
Artificial DNA transcription also just used ordinary RNA polymerase enzymes derived from natural organisms or bacteria ( this p.7 ), instead of using truely artificial enzymes.
Attempt to create some artificial enzymes mimicking catalytic sites of natural enzymes is limitted only to a few kinds of proteins like oxidase enzymes by the trial-and-error-discovery approach ( this p.1-left-last-paragraph, p.7 = antibody ) without looking into molecular mechanism (= still unknown, its side effect is unpredictable, so still useless for medical treatment, this p.18-right~p.19 ).
No quantum mechanics (= useless ) is used in this molecular biology ( this p.2-7 ).
One of those few artificial proteins studied now is PET hydrolase called FraC = pore-forming toxins obtained from the natural sea anemone, whose slightly-genetically-modified version by DNA mutagenesis ( this p.10-11, this p.44(or p.43) ) is expected to degrade plastics called PET, failed to degrade macro-PET plastics (= so useless artificial proteins ), and showed No improvement over other natural enzymes such as LCC enzymes obtained from plant-cell degrading microorganisms ( this-abstract-last, Fig.7, p.9-left-2nd-paragraph ).
↑ Scientists just repeat the same impractical time-consuming mainstream protein simulating methods consisting of molecular dynamics (= MD ) and one-pseudo-electron DFT approximation, which cannot predict anything, in vain with No progress ( this p.9-right, p.4-Fig.3h=time-consuming MD can simulate only 200ns-very-short reaction, which cannot simulate important longer biological reactions. this 2.materials and methods ).
The so-called DNA nanorobots or DNA origami are just multiple DNA strands sticking to each other based on complementary sequences by roughly applying heat or UV lights ( this p.3 ) without considering atomic interaction nor manipulating each atom, molecule, and No proteins are involved, so completely useless for drug development ( this p.4, this p.9 ).
The 2nd, 6th, 8th, last paragraphs of this hyped news say
"In synthetic biology, researchers try to recreate crucial mechanisms of life in vitro, such as cell division. The aim is to be able to synthesize minimal cells. The research team.. has now synthetically reproduced contractile rings for cell division using polymer rings composed of DNA nanotubes (= false, this research has nothing to do with cell division )."
"They have developed a theoretical description and a molecular dynamics (= MD ) simulation of the contraction mechanism, which match the experimental results of their research partners (= pseudo-classical molecular dynamics based on artificial potential or force fields has No ability to predict experimental results due to useless quantum mechanics, this p.5-lower )"
"In this way, it is possible to determine how the diameter of the DNA ring can be precisely controlled, which is highly significant for future (= just speculation, still useless now ) applications of contractile rings in synthetic biology."
"Mechanisms for cell division are an important step towards an artificial cell (= this research has nothing to do with artificial cell, though )"
↑ This research just combined DNA nanotubes with adhesive peptides called starPEG to make the ring of DNA nanotubes, and No cell division was involved, so this has nothing to do with artificial cell or cell division (= this is overhyped fake news ).
↑ This research paper ↓
p.2-Fig.1 shows DNA nanotubes (= brown ) joined together through the adhesive peptides (= blue ) called starPEG, and No cell or cell division was involved in this process (= only DNA tubes and peptides were used ).
p.3-right-last-paragraph says "We find that starPEG-(KA7)4 promotes the
efficient formation of DNA nanotube rings with several micrometers in
diameter"
p.4-Fig.3 DNA nanotubes and the adhesive peptides starPEG alone formed DNA ring without cells. ← No cell division was involved. p.5-Fig.4, p.10-Molecular dynamics simulation (= coarse-grained ) treated DNA nanotubes as fictitious beads, each bead diameter is σ = about 12nm
↑ To simulate the DNA nanotube ring formation, they relied on (pseudo-classical) coarse-grained molecular dynamics (= MD ) where DNA nanotubes were treated as a collection of fictional big beads (= each bead's interaction size is about 12nm, but it has No shape ), and the interaction parameters (= ε ) of the fictional potential between DNA beads were artificially-adjusted to fit the observed ring size ( this p.18-19, this p.6-Fig.5c ).
No detailed atomic information (= instead, much bigger fake bead model was used for roughly approximating DNAs ), No cell divition, No peptides, No solvent molecules were considered in this fictional coarse-grained molecular dynamical (= MD, this p.10-molecular dynamical ) simulation which is useless, cannot clarify the detailed mechanism of DNA nanotubes at atomic level, much less cell division.
↑ If they used the normal molecular dynamical simulation (= all-atom ) distinguishing individual atoms, each time step of computing and updating each atomic position must be less than 2 femtoseconds (= 2fm ), which must repeat 109 calculation steps (= take unrealistically too much time ! ) to simulate 1 μs molecular motions that cannot simulate important biological reactions with seconds ~ hours time scales.
In this research, they used much bigger fictional beads instead of actual smaller atoms sacrificing atomic-level accuracy, so each time step could be longer = 0.02 ns (= instead of 2fs ), and the simulation time was also longer = 20ms (= after repeating 109 time steps, this p.21-lower ), but even this fictional DNA bead models took too much time to simulate actual DNA nanotube ring formation that would take several seconds ~ minutes.
So they artificially changed parameters and used fake viscosity η ( this p.22-upper ) to reproduce the DNA nanotube ring formation by force.
This-middle-Goal says
"However, this coarse-graining of time in molecular simulations is only poorly understood,.. While one can resort to approximate techniques, such as the introduction of an ad hoc speed-up factor,.. the applicability of this rule of thumb is necessarily limited and Not based on physical insights (= coarse-grained or CG MD often failed, this p.2-left-lower )."
↑ This research just simulated meaningless DNA nanotubes' ring formation expressed as fictional bead model, did Not simulate any cell division or protein interaction, so No practical application.
Quantum mechanical unphysical models cannot give real atomic shapes, so physicists have to rely on artificially-created (pseudo-)potentials, and extremely-time-consuming molecular dynamical simulation to describe molecular or protein motions that are unable to simulate actual cell's functions at precise atomic levels. ← No effective drug discovery.
The so-called quantum Biology is just a useless overhyped pseudo-science irrelevant to real biology ( this-lower-quantum biology today, this last-paragraph, this-lower Looking ahead, this 4-5th-paragraphs, this 6th-paragraph ).
Quantum biology (= still in its infancy, impractical forever, this 2nd-last paragraph, this p.2-left-1st-paragraph ) tries to unscientifically explain photosynthesis using the baseless imaginary quasi-particle called exciton ( this 3-5th paragraphs, this p.6-right-lower ) or deceptively-vague quantum effects in vain ( this p.11-right-2nd-paragraph, this p.7-second-paragraph, this 7th-paragraph, this p.3-scalable fractality ).
The current human technology cannot design or manufacture truly-artificial proteins or enzymes for curing diseases due to useless atomic physics or fictional quantum mechanics.
CRISPR-Cas9 gene-editing system was also discovered and extracted from natural bacterial immune system, Not designed nor created by human's technology stalled by quantum mechanics.
Due to the useless mainstream atomic theory such as quantum mechanics, DFT, molecular dynamics, biologists unable to design or create truly useful artificial proteins from atomic theory have to rely on slight chance that they may luckily find some natural enzymes or antibodies obtained from immunizing natural animals (or xenotransplanted mouse ) usable for treating cancers (= called immunotherapy ), which attempts often failed.
CRISPR system obtained from natural bacteria is one of those enzymes researchers pin their hope on for future cancer treatment, but they still can Not find effective way to deliver CRISPR-cas9 enzymes to cancer cells inside human bodies, and they cannot prevent possible side effects where CRISPR may injure normal cells ( this p.11 ), which is far from effective treatment ( this 10th-paragraph, this p.14-conclusion ).
The recent research paper and hyped news fruitlessly try to explain some mechanism of this CRISPR enzyme using the impractical old quantum mechanical DFT or Kohn-Sham theory (+ meaningless AI ) which replaces actual many-electron molecules by only one fictitious pseudo-electron ( this p.2-2nd-paragraph ) or quasi-particle ( this p.10 ) with artificially-chosen fictitious potentials (= unknown functionals, this p.6, this p.2-right-2nd-paragraph ) and adjustable empirical parameters ( DFTB with empirically-fitted ad-hoc parameters in this Materials and methods-Feature matrix, this p.21 ).
↑ This unphysical mainstream quantum mechanical DFT with one pseudo-electron model is unable to describe actual biological system in a practical realistic way, and unable to predict any physical values from the original useless quantum mechanics ( this p.15 ), which is why (quantum) biology stalls with No utility for drug development.
All the curent biological and medical researches are useless, just a waste of time only in aiming at publishing papers in (top) journals, instead of really aiming at curing diseases.
Because the mainstream atomic theory quantum mechanics and its most pupular density functional theory (= DFT ) lack real atomic picture with real shapes or masses, which unreal quantum model leads to unrealisically-time-consuming molecular dynamics (= MD ) as the only choice in molecular simulating methods, which prevent all the applied science from advancing.
↑ To hide this inconvenient fact of the stalled science and innovation, academia, corporations and the media are colluding to create and spread overhyped exaggerated news every day.
The last paragraph of this hyped news says
"The outcomes of this research will (= just speculative future, still realized nothing ) enable the design of new catalysts for the efficient and sustainable production of green hydrogen. Work is already underway (= still unrealized ) at the .. (?)"
↑ This research just tried to explain some photocatalysis by using impractical one-pseudo-electron DFT model with artificially-fitting parameters U, chosen exchange functionals with No realistic atomic picture nor quantum mechanical prediction.
↑ this p.8-one-pseudo-electron DFT with artificially-chosen PBE exchange functional + freely-adjustable U parameters combined with another empirical HSE06 functional ( this p.3-left-(4) ) with adjustable Fermi spearing parameter.
this p.6-effective DFT by adjusting the width of the corresponding Fermi smearing, and U parameter (= No quantum mechanical prediction nor real atomic picture )
The last paragraph of this hyped news says
"This drug-free, target-specific, and biologically benign nanozyme has great potential (= this word means just baseless speculation, still realized nothing ) as a potent biocatalyst for use in safe cancer treatment".
↑ This research also tries to explain some enzyme by using impractical one-pseudo-electron DFT model with freely-chosen exchange functional and parameters = No quantum mechanical prediction ( this 3rd-paragraph ).
The 2nd-last and last paragraphs of this hyped news say
"The new method could (= still speculation ) in principle be harnessed to produce both natural molecules and nonnatural molecules.
Whether these things will (= uncertain future, still useless ) ever be done is hard to say.."
↑ This research tried to explain some chemical reactions using one-pseudo-electron DFT model with freely-chosen empirical exchange functional with No quantum mechanical prediction ( this p.3-right-last-DFT-empirical ωB97X-D4 functional this p.5-right, this p.54-7-Grimme's dispersion DFT-D3-artificially-adjustable damping parameters )
The 3rd and last paragraphs of this hyped news say
"In particular, quasiparticles (= unreal useless model ) that are part light (photons) and part matter (electrons and lattice vibrations), called surface plasmon-phonon polaritons, are formed at the rippled stack interplaying strongly with the molecules atop."
"We are confident that this study will (= uncertain future, still useless ) contribute to the development of new lab-on-chip devices (← ? )"
↑ This experiment tried to explain some interaction of light and molecule using fictitious quasiparticle models such as plasmon (= expressed as just nonphysical math operators with No real shape, this p.4-left-upper ) and phonon with freely-adjustable parameters ( this Fig.6 and lower ) with No quantum mechanical prediction.

2023/12/10 updated. Feel free to link to this site.